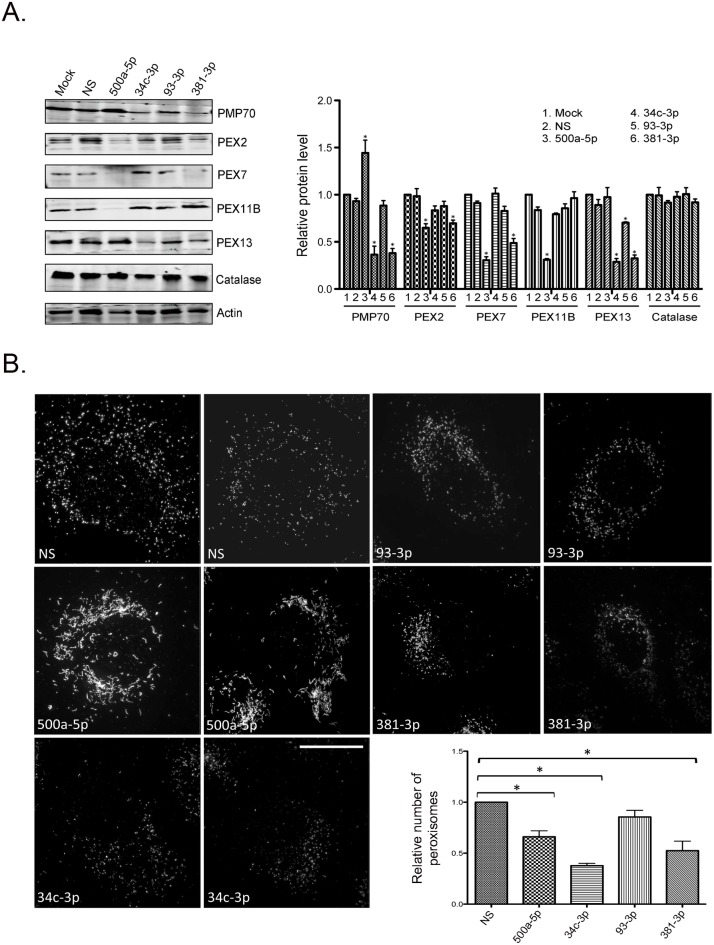
Fig 3. A subset of HAND-associated miRNAs reduces expression of peroxisomal proteins and alters peroxisome abundance and/or morphology.
A. A549 cells were transfected with mimics (30 nM) for miR-NS, miR-500a-5p, miR-34c-3p, miR-93-3p or miR-381-3p. Forty-eight hours later, cell lysates were subjected to immunoblot analyses. PMP70 and actin were detected using primary mouse monoclonal antibodies and secondary donkey anti-mouse IgG conjugated to Alexa Fluor 680. PEX2, PEX7, PEX11B, PEX13 and catalase were detected using primary rabbit antibodies and secondary goat anti-rabbit IgG conjugated to Alexa Fluor 680. Relative peroxisomal protein levels (normalized to actin) in mock- and miRNA-transfected cells from three independent experiments are shown. Bars represent standard error of the mean. B. A549 cells were transfected with 30 nM of mimics for miR-NS, miR-500a-5p, miR-34c-3p, miR-93-3p or miR-381-3p for 38 hours after which they were processed for super resolution microscopy. Peroxisomes were identified using a mouse monoclonal antibody to PMP70 and donkey anti-mouse IgG conjugated to Alexa Fluor 488. Nuclei were stained with DAPI. Images were acquired and reconstructed using a DeltaVision OMX structured illumination microscope. Size bar is 10 μM. The relative numbers of peroxisomes in cells transfected with each miRNA were determined using Volocity image analyses software from three independent experiments (minimum of 20 cells). Bars represent standard error of the mean. *, p<0.05.