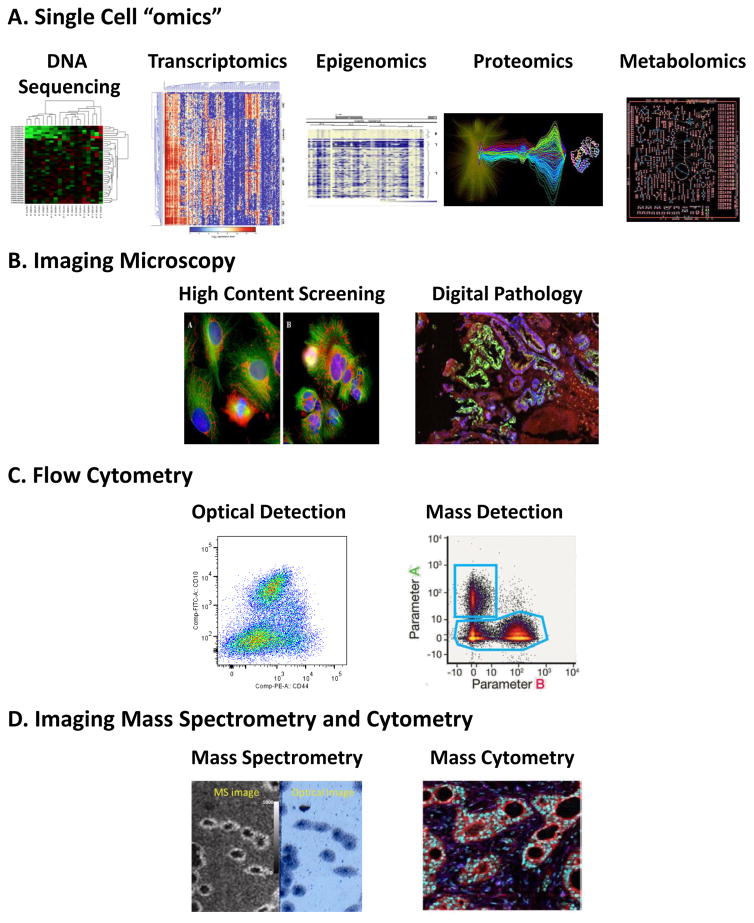
Figure 3.
Heterogeneity in populations of cells can be quantified by a variety of methods that permit cell-by-cell measurements. A. Single cell genomics, epigenomics, proteomics and metabolomics (reprinted with permission from Spagnolo et al.21), and/or transcriptomics (from Saadatpour et al.235) to study heterogeneity use either ground up tissue samples or single cells and can provide a comprehensive analysis of heterogeneity for a large number of cells. B. High Content Screening and Digital Pathology21 employs multiple fluorescent probes to capture a broad range of information including expression levels and subcellular localization of molecules within and across individual cells. C. Optical (from Hines et al.114) and mass cytometry (from Spitzer and Nolan105) can provide information on expression levels of several molecules simultaneously as well as some morphological information in large populations but do not report spatial heterogeneity. D. Mass Spectrometry readouts expand the range of molecules that can be simultaneously detected in flow cytometry (mass cytometry) and can be used to image tissues and cells in imaging Mass Cytometry (from Giesen et al.93) and Imaging Mass Spectrometry (from Zavalin et al.100).