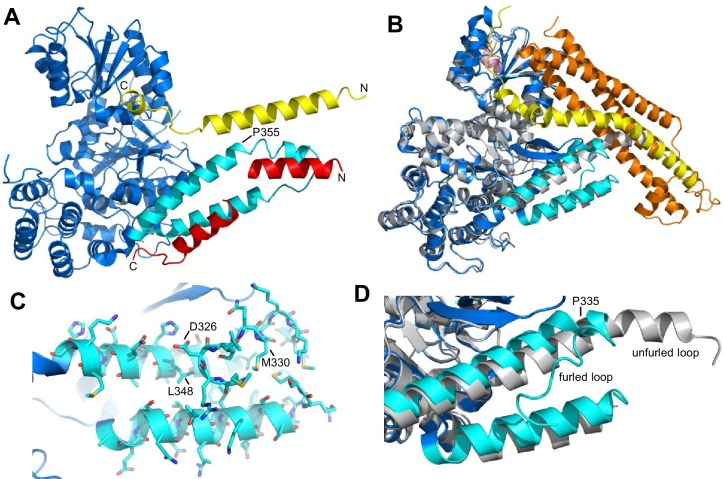
Figure 1. Autoinhibition by a loop of Munc18-1 is likely to inhibit binding to synaptobrevin.
(A) Ribbon diagram of the structure of Vps33 (blue) bound to the Nyv1 SNARE motif (red) (PDB code 5BV0) superimposed with the structure of Vps33 (not shown) bound to the Vam3 SNARE motif (yellow) (PDB code 5BUZ) (Baker et al., 2015). The two helices connected by a loop in domain 3a of Vps33, which are involved in Nyv1 binding, are shown in cyan. (B) Crystal structure of Munc18-1 (blue) bound to closed syntaxin-1 (SNARE motif in yellow; N-terminal region in orange) (PDB code 3C98) (Burkhardt et al., 2008) superimposed to the crystal structure of Munc18-1 (gray) bound to a syntaxin-4 N-terminal peptide (pink) (PDB code 3PUJ) (Hu et al., 2011). The helix-loop-helix of domain 3a of Munc18-1 in the complex with syntaxin-1 is in cyan. (C) Close-up of the helix-loop-helix of domain 3a of Munc18-1 bound to syntaxin-1 (not shown) illustrating how the loop forms a ‘furled conformation’ by folding back onto the groove between the two helices that is putatively involved in synaptobrevin binding. Side chains are shown as stick models and those that were mutated to disrupt synaptobrevin binding are labeled. (D) Close-up of the helix-loop-helix region from the superposition shown in (B) but without showing syntaxin-1. Note how in the complex with closed syntaxin-1, the Munc18-1 loop is furled (cyan). whereas in the complex with the syntaxin-4 N-terminal peptide, the loop is unfurled and forms a helical extension of one of the helices but with a bend (gray). This bend has P335 in the corner, and the homologous residue of Vps33 (P355) is also in the corner of the helix bend (A).