Abstract
EGFR (exon 19 and exon 21) mutations in patients with advanced non-small cell lung cancer (NSCLC) treated by EGFR-TKIs are associated with a better survival; while KRAS mutations predict a worse prognosis. However, there are divergent findings regarding the prognostic value of EGFR and KRAS mutations in circulating tumor DNA (ctDNA). We aimed to summarize the evidence for the use of circulating EGFR and KRAS mutations as prognostic factors in advanced NSCLC patients.
We searched the network databases for studies reporting progression-free survival (PFS) and overall survival (OS) stratified by EGFR or KRAS mutations in ctDNA in advanced NSCLC patients. Thirteen studies enrolling 2,293 patients were reviewed. Correlation of circulating EGFR or KRAS mutations with patients’ prognosis was assessed by meta-analysis.
The pooled analyses showed that EGFR mutations in ctDNA significantly prolong PFS (HR=0.64,95% CI 0.51-0.81, I2=0%, p=0.0002), namely, in patients treated by EGFR-TKIs. There is a trend to have a prolonged OS for advanced NSCLC patients with circulating EGFR mutations who were treated by EGFR-TKIs (HR=0.79, 95% CI 0.52-1.21, I2=0, p=0.28). KRAS mutations detected in ctDNA predict a worse PFS (HR=1.83, 95% CI 1.40-2.40, p<0.0001) and OS (HR=2.07, 95% CI 1.54-2.78, p<0.00001) in advanced NSCLC patients treated by chemotherapy. Sensitivity analyses and subgroup analyses demonstrated the stability of our conclusion.
Our analysis showed that EGFR mutations in ctDNA predicted a better PFS, in particular in advanced NSCLC patients treated by EGFR-TKIs. KRAS mutations in ctDNA indicated a worse PFS and OS in patients treated by chemotherapy.
Keywords: EGFR, KRAS, NSCLC, prognosis, meta-analysis
INTRODUCTION
Non-small cell lung cancer (NSCLC) remains the major cause of cancer-related mortality. Studies showed that epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs) confer better outcome in patients with EGFR mutations (exon 19 deletions, exon 21 L858R point mutations) than in those with the wild type [1]. About 5-15% of NSCLC patients harbor EGFR mutations [2]. KRAS mutations predict worse prognosis among NSCLC patients treated by EGFR-TKIs or chemotherapy [3, 4]. KRAS mutations are detected in about 30% of NSCLC in white people [5]. Approximately 97% of KRAS mutations in NSCLC involve codon 12 or codon 13 [3, 6]. Several studies performed systematic review and meta-analysis to assess the prognostic value of EGFR and KRAS mutations in tumor tissue in NSCLC patients [4, 7–9].
Circulating tumor DNA (CtDNA) is shed into the bloodstream by tumor cells [10]. Evidence shows that ctDNA might be used as a noninvasive blood biomarker in tumor medicine [11, 12]. Diagnostic tests for ctDNA such as OncoBEAM® RAS CRC Kit (Sysmex Inostics GmbH), cobas® EGFR Mutation Test V2 (Roche) and EGFR Mutations Detection Kit (AmoyDx) are commercially available for ctDNA detection. The published papers offered divergent findings regarding the prognostic value of EGFR and KRAS mutations in ctDNA in patients with advanced NSCLC. Only one study by Mao et al. conducted meta-analysis to explore the prognostic value of EGFR in ctDNA in advanced NSCLC patients [13]. However, overlapping studies were included in their study [13]. No studies did a systematic review and meta-analysis to assess the prognostic value of KRAS mutations in ctDNA in patients with advanced NSCLC. Thus, we conducted a systematic review and meta-analysis to explore their prognostic values in advanced NSCLC patients.
RESULTS
Included studies
A total of 2,295 potential studies were identified. After screening by title and abstract, 2,216 studies were excluded. The main reasons for exclusion were duplicative studies, reviews, not human studies, not relevant to ctDNA, incorrect tumor type and epigenetic alterations. Of the remaining 79 studies, the full text was screened and 66 studies were excluded for lack of follow-up, no information about prognosis, not restricted to advanced NSCLC patients, non-English literature, not restricted to KRAS or EGFR (exon19 and exon21) mutations. Finally, 13 studies met the inclusion criteria and were included for systematic review and meta-analysis (Figure 1).
Figure 1. PRISMA 2009 Flow Diagram.
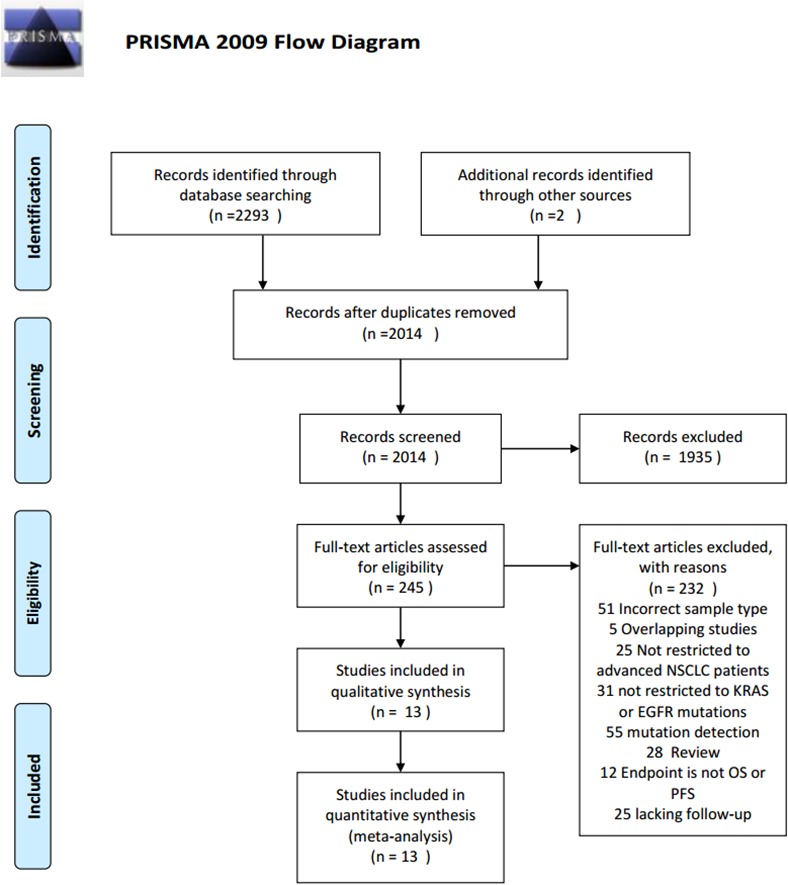
PRISMA flow diagram for study selection.
Study characteristics
Thirteen studies containing 1,452 patients were published between 2005 and 2013. These studies analyzed the relationship between circulating EGFR and KRAS mutations status and survival outcomes. The mean number of patients for each study was 67, ranging from 22 to 308. Four studies were retrospective and 9 were prospective. All studies were published in full and all had enough information to obtain the hazard ratios (HRs) and associated 95% confidence intervals (CIs). The main characteristics of the included publications are summarized in Table 1.
Table 1. General characteristics of the study populations.
| First author name (year) | Country | Publication type | Study design | Patients included in analysis | Male sex N (%) | Median age, y/o | Never smoker/ total | Tumor stage | Treatment | Detection matrix | Sampling time | ctDNA detection | Mutation detection methods | ctDNA positive, n |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mack 2009 | USA | Full publication | Pro | 45 | NR | NR | NR | ad | TKIs | Plasma | Prior to treatment | EGFR/KRAS | DS | 6EGFR, 2KRAS |
| Camps 2005 | Spain | Full publication | Pro | 67 | 94 | 64 | 7 | ad | Chemo | Serum | Prior to treatment | KRAS | DS | 20 |
| Camps 2011 | Spain | Full publication | retro | 308 | 83.8 | 60 | NR | ad | Chemo | Plasma | Prior to treatment | KRAS | Real-time PCR | 27 |
| Huang 2012 | China | Full publication | Pro | 207 | NR | NR | 46.4 | ad | TKIs | Plasma | Prior to treatment | EGFR | DHPLC | 70 |
| Xu 2012 | China | Full publication | Pro | 51 | 60.7 | 54 | 62.7 | ad | gefitinib | Plasma | Prior to treatment | EGFR | ME- Liquidchip |
15 |
| Kimura 2007 | Japan | Full publication | Retro | 42 | 66.7 | 58 | 33.3 | ad | gefitinib | Serum | Prior to treatment | EGFR | SARM | 7 |
| Kim 2013 | South Korea | Full publication | Pro | 22 | NR | NR | NR | ad | TKIs | Serum | Prior to treatment | EGFR/KRAS | EGFR: PNA-LNA; KRAS: DS | 4KRAS ; 5EGFR |
| Bai 2009 | China | Full publication | Pro | 102 | NR | NR | NR | ad | gefitinib | Plasma | Prior to treatment | EGFR | DHPLC | 37 |
| Punnoose 2012 | USA/ Australia | Full publication | Retro | 37 | 56.76 | NR | 19.35 | ad | erlotinib/pertuzumab | Plasma | Prior to and during treatment | KRAS/EGFR | SARMS | 4EGFR, 5KRAS |
| Zhuo 2011 | China | Full publication | Retro | 145 | 59 | M:60.5, W: 62 | NR | ad | chemo | Plasma | Prior to treatment | EGFR | DHPLC | 54 |
| Qin 2011 | China | Full publication | Pro | 46 | NR | NR | NR | ad | gefitinib | Plasma | NR | EGFR | SARMS | 18 |
| Nygaard 2013 | Denmark | Full publication | Pro | 246 | 61 | 66 | NR | ad | chemo | Plasma | Prior to treatment | KRAS | In-house real-time PCR | 43 |
| He 2009 | China | Full publication | Pro | 134 | 6.4 | 60 | NR | ad | surgery/ chemo/ TKIs | Plasma | Prior to treatment | EGFR | DS | 66 |
Abbreviations: Pro: prospective study; Retro: retrospective study; NR: no report; chemo: chemotherapy; ad: advanced stage; DS: direct sequencing; SARMS: Scorpion Amplification Refractory Mutation System; DHPLC: denaturing high-performance liquid chromatography; PNA-LNA: peptide nucleic acid-locked nucleic acid; ME-liquid chip : mutant-enriched liquid chip
QUALITY ASSESSMENT
We assessed risk of bias using the Cochrane Collaboration's tool (the Cochrane Collaboration's tool for assessing risk of bias in randomized trials) [14]. All the included studies had a low risk of bias, as summarized in Figure 2.
Figure 2. Our judgements about each risk of bias item of included studies.
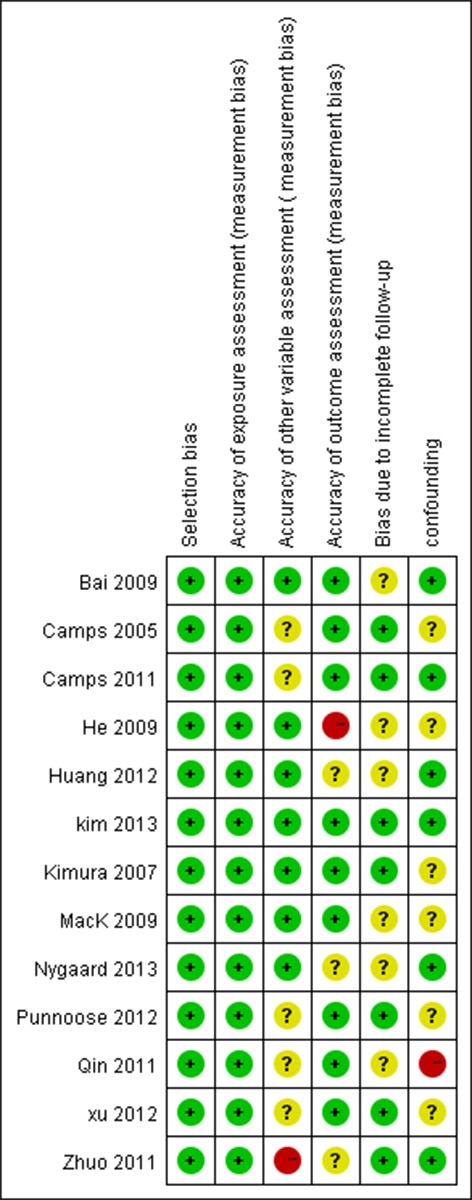
Red circles represent studies with high risk of bias; Green circles represent studies with low risk of bias, yellow circles represent studies with uncertain risk of bias.
EGFR (exon 19 and exon 21) mutations and prognosis
Relationship between EGFR mutations (exon 19 and exon 21) and PFS
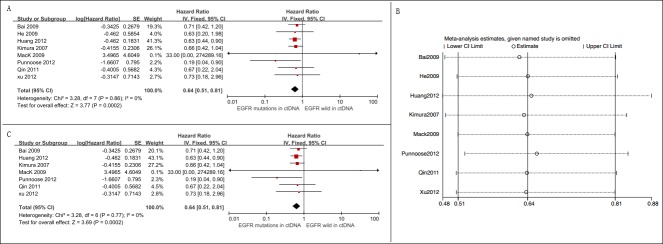
Eight studies assessing the relationship between EGFR mutation status in ctDNA and PFS were eligible for the meta-analysis [15–22]. A total of 705 patients were included, and 248 were EGFR mutation-positive. Among them, 684 patients (97%) were treated by TKIs, and the rest 21 (3%) were treated by chemotherapy. The overall summary HR was 0.64 (95% CI 0.51-0.81), with a low degree of heterogeneity (p = 0.86, I2 = 0%). The pooled analysis indicated a better PFS for circulating EGFR mutation-positive patients (Figure 3A).
Figure 3. Meta-analysis of the prognosis of circulating EGFR mutations for PFS.
A. Forest plots of HR and 95% CI in advanced NSCLC patients. Patients with circulating EGFR mutations had a better PFS (HR = 0.64, 95% CI 0.51-0.81); B. results of sensitive analysis showed that there was no “dominant” study driving the results of the meta-analysis; C. forest plots of HR and 95% CI in advanced NSCLC patients treated by EGFR-TKIs. Circulating EGFR mutations indicated a better PFS among patients who were treated by EGFR-TKIs (HR = 0.64, 95% CI 0.51-0.81).
Sensitive analysis
Sensitivity analysis by “leave-one-out” strategy showed that there was no “dominant” study driving the results of meta-analysis (Figure 3B).
Six of 8 studies enrolled Asian patients [15, 17, 18, 20–22], one study included both Asian and Western patients [16], and rest one included only Western patients [19]. We changed our criteria by including studies with Asian patients only. The aggregated analysis showed that EGFR mutations prolonged PFS among Asian patients (HR = 0.66, 95% CI 0.52-0.83, I2 = 0%) (Supplementary Figure S1).
Seven studies examined the relationship between EGFR mutations status and PFS among advanced NSCLC patients with TKIs therapy [15–19, 21, 22]. The overall summary HR was 0.64 (95% CI 0.51-0.81, I2 = 0%), suggesting that the observed benefit is tightly linked to the effect of EGFR-TKIs in the setting of EGFR mutated patients (Figure 3C).
Subgroup analysis
We performed subgroup analyses on the basis of detection matrix (serum vs. plasma) and study year (prior to 2010 vs. after 2010). There was no statistical significance between these subgroups (Table 2).
Table 2. Subgroup analyses on the basis of detection matrix and study year for EGFR mutations and progression-free survival analysis.
| Trial characteristic | Subgroup analysis | |||||
|---|---|---|---|---|---|---|
| Stratification variable | Number of study arms | Pooled hazard ratios | 95% CI | p-value within subgroups | p-value between subgroups | |
| Detection matrix | Serum | 1 | 0.66 | 0.42–1.04 | 0.07 | 0.89 |
| Plasma | 7 | 0.64 | 0.51–0.81 | 0.78 | ||
| Study year | Before 2010 | 4 | 0.68 | 0.49–0.94 | 0.86 | 0.62 |
| After 2010 | 4 | 0.61 | 0.44–0.84 | 0.52 | ||
Relationship between EGFR (exon 19 and exon 21) mutations and overall survival
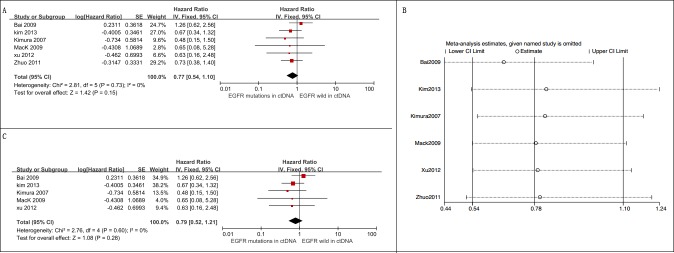
Six studies with 407 patients examining the relationship between circulating EGFR mutation status and OS among advanced NSCLC patients were included [15, 19, 21–24]. Among them, 124 were circulating EGFR mutation-positive. Our pooled analysis showed that there is a trend for longer OS in patients harboring circulating EGFR mutations (HR = 0.77, 95% CI 0.54-1.10, I2 = 0%) (Figure 4A).
Figure 4. Meta-analysis of the prognosis of circulating EGFR mutations for OS.
A. Forest plots of HR and 95% CI in advanced NSCLC patients. Only marginally statistically significant OS (HR = 0.77, 95% CI 0.54-1.10) was observed between NSCLC patients with and without circulating EGFR mutations; B. results of sensitive analysis showed that there was no “dominant” study driving the results of the meta-analysis; C. forest plots of HR and 95% CI in advanced NSCLC patients treated by EGFR-TKIs, there was no statistical significance between patients with and without circulating EGFR mutations (HR = 0.79, 95% CI 0.52-1.21, p = 0.15).
Sensitive analysis
The “leave-one-out” strategy demonstrated that there was no dominant study driving the results of meta-analysis (Figure 4B).
Five of 6 studies enrolled patients from Asia [15, 21–23, 25]; only 1 study included patients from the West [19]. We changed our criteria by including studies with Asian patients. This change did not alter our conclusion (HR = 0.78, 95% CI 0.54-1.11, I2 = 0%) (Supplementary Figure S2).
Five studies reported the relationship between EGFR mutation status and OS among advanced NSCLC patients treated by TKIs [15, 19, 21, 22, 25]. The pooled analysis revealed that in the setting of EGFR-TKIs treatment, EGFR-mutation positive patients trend to have a longer OS (HR = 0.79, 95% CI 0.52-1.21, I2 = 0) (Figure 4C).
Subgroup analysis
Subgroup analyses on the basis of detection matrix (serum vs. plasma) and study year (prior to 2010 vs. after 2010) were performed. There was no statistical significance between these subgroups (Table 3).
Table 3. Subgroup analyses on the basis of detection matrix and study year for EGFR mutation and overall survival analysis.
| Trial characteristic | Subgroup analysis | |||||
|---|---|---|---|---|---|---|
| Stratification variable | Number of study arms | Pooled hazard ratios | 95% CI | p-value within subgroups | p-value between subgroups | |
| Detection matrix | Serum | 2 | 0.61 | 0.34–1.10 | 0.62 | 0.33 |
| Plasma | 4 | 5.13 | 2.43–10.82 | 0.66 | ||
| Study year | Before 2010 | 3 | 0.93 | 0.52–1.67 | 0.35 | 0.42 |
| After 2010 | 3 | 0.69 | 0.44–1.08 | 0.97 | ||
KRAS mutations and prognosis
Relationship between KRAS mutations in ctDNA and PFS
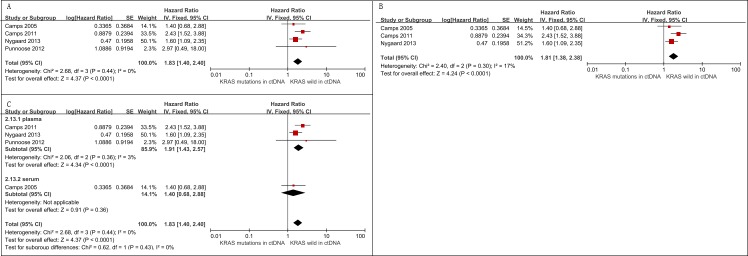
Four studies with 658 patients assessing the relationship between KRAS mutations in ctDNA and PFS among advanced NSCLC patients were eligible for meta-analysis [3, 6, 16, 24]. Among them, 95 were circulating KRAS mutation-positive. In this analysis, KRAS mutations in ctDNA were associated with a worse PFS (HR = 1.83, 95% CI 1.40-2.40, I2 = 0) (Figure 5A).
Figure 5. Meta-analysis of the prognosis of circulating KRAS mutations for PFS.
A. Forest plots of HR and 95% CI in advanced NSCLC patients. The pooled analysis showed that circulating KRAS mutations were associated with a worse PFS (HR = 1.83, 95% CI 1.40-2.40); B. forest plots of HR and 95% CI in advanced NSCLC patients treated with chemotherapy. Circulating KRAS mutations were associated with a shorter PFS among patients treated by chemotherapy (HR = 1.81, 95% CI 1.38-2.38); C. Subgroup analyses on the basis of detection matrix (serum vs plasma) indicates no statistical significance between these KRAS mutations detected in serum and KRAS mutations detected in plasma regarding PFS (p = 0.43).
Sensitive analysis
Three out of 4 studies explored KRAS mutations in ctDNA as a predictive factor for the effect of chemotherapy on advanced NSCLC patients [3, 6, 24]. The pooled analysis showed that KRAS mutations in ctDNA indicate a worse PFS in patients treated by chemotherapy (HR = 1.81, 95% CI 1.38-2.38, I2 = 17%) (Figure 5B).
The “leave-one-out” strategy showed that there was no dominant study driving the results of meta-analysis (Supplementary Figure S3)
Subgroup analysis
We performed subgroup analysis based on detection sample (serum vs. plasma). There was no statistical significance between these subgroups (Figure 5C).
Relationship between KRAS mutations in ctDNA and OS
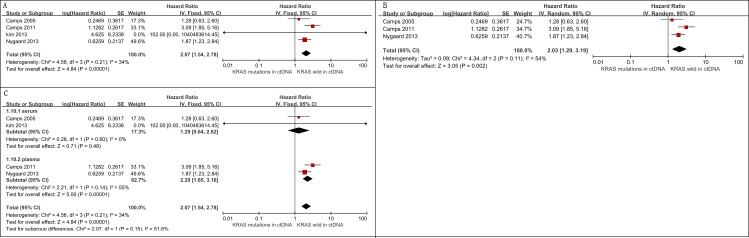
Five studies assessed the relationship between KRAS mutation status and OS among advanced NSCLC patients [3, 6, 24–26]. A total of 693 patients were included and 106 were KRAS mutation-positive. Findings from the meta-analysis suggested that KRAS mutations in ctDNA were associated with an unfavorable OS (HR = 2.07, 95% CI 1.54-2.78, I2 = 34%) (Figure 6A).
Figure 6. Meta-analysis of the prognosis of circulating KRAS mutations for OS.
A. Forest plots of HR and 95% CI in advanced NSCLC patients; patients with circulating KRAS mutations had a worse OS (HR = 2.07, 95% CI 1.54-2.78) B. forest plots of HR and 95% CI in advanced NSCLC patients treated with chemotherapy; circulating KRAS mutations were associated with a worse OS among patients treated by chemotherapy (HR = 2.03, 95% CI 1.29-3.19) C. subgroup analysis in advanced NSCLC patients on the basis of detecting matrix (serum vs. plasma). The subgroup difference was not significant (p = 0.43).
Sensitive analysis
Three of 4 studies examined KRAS mutations in ctDNA as a predictive factor among advanced NSCLC patients who were treated with chemotherapy [3, 6, 24]. Our pooled analysis showed that patients with KRAS mutations in ctDNA had a worse OS (HR = 2.03, 95% CI 1.29-3.19, I2 = 54%) (Figure 6B).
The “leave-one-out” sensitivity analysis showed that our results were stable (Supplementary Figure S4).
Subgroup analysis
We performed subgroup analysis based on detection sample (serum vs. plasma). The pooled analysis demonstrated that the association between OS and KRAS mutation status was slightly stronger in plasma (HR = 2.29, 95% CI 1.65-3.16) than in serum (HR = 1.29, 95% CI 0.64-2.62), though the subgroup difference was not significant (p = 0.15) (Figure 6C).
DISCUSSION
EGFR genotyping has become a routine test for the selection of patients as candidates for TKIs therapy. KRAS mutations test may help to predict less benefit of treatment with EGFR-TKIs and chemotherapy.
EGFR and KRAS mutation tests are often carried out from formalin-fixed paraffin embedded (FFPE) tumor tissue samples. However, molecular testing with FFPE has some limits. DNA extracted from FFPE is fragmented and also contains DNA lesions (including uracil and thymine deriving from cytosine deamination) that may lead to sequence artifacts [27]. Frozen tumor biopsies and fresh biopsies may complement FFPE. However, sometimes this is not feasible in clinical practice for advanced NSCLC, especially for successive dynamic monitoring. Tumor heterogeneity is another hurdle when utilizing tissue samples for mutation tests [28]. CtDNA may overcome these limits, and may be used as a surrogate [29]. However, we still have a limited understanding of the origination of ctDNA. If the ctDNA analyzed originates from apoptotic/necrotic tumor cells, it may have limited applicability for prognostic analysis. If the ctDNA utilized is from actively shedding tumor cells (either in circulation or in primary/metastatic sites), it may provide valuable information for treatment decision and prognostic prediction. Due to tumor heterogeneity, the choice of target treatment should ideally be based on the ctDNA from metastatic sites instead of on that from the primary sites [30]. Therefore, the limited understanding of its origination arises the question whether EGFR or KRAS mutations detected in ctDNA could be used as biomarkers for prognosis prediction.
The current meta-analysis established that advanced NSCLC patients with ctDNA EGFR mutations predicted a better PFS, namely, in patients treated by EGFR-TKIs. There is a trend to have a prolonged OS for advanced NSCLC patients with ctDNA EGFR mutations who were treated by EGFR-TKIs. Mao et al. carried out meta-analysis in advanced NSCLC patients treated by TKIs and arrived at a different conclusion. Their analyses showed that EGFR mutations detected in blood were associated with better OS (HR = 0.71, 95% CI 0.50-0.99, p = 0.61) [13]. The difference was likely to be caused by including suspected overlapping studies in that study [13]. Moreover, most of the HR estimates were extrapolated from the survival curves, which also contributed to this discrepancy. Huang et al. and Lee et al. carried out meta-analyses in NSCLC patients with tissue EGFR mutations. They found that the improvement in OS was only marginally statistically significant in patients receiving TKIs therapy, which was similar to our conclusion [7, 8].
Our meta-analysis showed that at least in patients treated by chemotherapy, circulating KRAS mutations correlated with worse PFS and OS. This finding was consistent with the meta-analysis conducted by Chen et al, who evaluated the prognostic value of tissue KRAS mutation status [4].
Either serum or plasma was used as detecting matrix for genotyping in the eligible studies. The amounts of cell-free DNA are much higher in serum due to cell lysis during sample processing, hence reducing the fraction of tumor DNA in serum. However, this discrepancy did not alter our conclusions.
This meta-analysis had some advantages. First, we performed a comprehensive review and reported the most up-to-date published data. Second, no heterogeneity was found in this meta-analysis. Finally, this was the first meta-analysis to assess the prognostic value of circulating KRAS mutations in advanced NSCLC patients.
Despite our efforts to provide an accurate and comprehensive analysis, the limitations of our meta-analysis should be highlighted. First, EGFR exon 19 and exon 21 respond differently to TKIs treatment [9]; thus, it is necessary to perform subgroup analysis according to EGFR mutation subtypes in a future study. Second, we did not perform subgroup analyses based on age, sex, smoking status, and detection methods due to insufficient data. Another limitation of our meta-analysis is that the status of other actionable mutations such as ALK rearrangements were not considered and should be included in an analysis with more available data in future studies. Finally, the eligible studies only performed univariate analyses; we cannot infer from our meta-analysis whether EGFR or KRAS mutations in ctDNA could be an independent factor or not.
Selection bias may exist in our paper. For EGFR and KRAS involved in lung cancers, striking differences in molecular alterations of these genes have been found in never and ever smokers [31]. Epidemiological studies of lung cancer showed that EGFR mutations occur more frequently in never smoker East Asia, while KRAS mutations occur more frequently in smokers and less common in never smoker East Asia [30]. In our paper, the rate of EGFR mutations is higher than usually expected, and the rate of KRAS mutations is much lower in the selected studies. This may be due to a high rate of never-smoker patients in our study.
Despite the aforementioned limitations, this meta-analysis suggested that EGFR mutations detected in ctDNA were associated with a better PFS, namely, in patients treated by EGFR-TIKs. There is a trend to have a prolonged OS for patients with ctDNA EGFR mutations who were treated by EGFR-TKIs. Circulating KRAS mutation-positive status in advanced NSCLC predicts a worse PFS and OS in patients treated by chemotherapy.
MATERIALS AND METHODS
Search methods for identification of studies
Meta-analysis Of Observational Studies in Epidemiology (MOOSE) guidelines were conformed to identify potential relevant studies. We did systematic electronic searches of Medline, Embase, Web of Science, the Cochrane Library, and Scopus up to October 10, 2015 (no start date limit was applied). The search strategy used was as follows: “Carcinoma, Non Small Cell Lung”, “Carcinomas, Non-Small-Cell Lung”, “Lung Carcinoma, Non-Small-Cell”, “Lung Carcinomas, Non-Small-Cell”, “Non-Small-Cell Lung Carcinomas”, “Non small Cell Lung Cancer”, “Non-Small-Cell Lung Carcinoma”, “Non Small Cell Lung Carcinoma”, “Carcinoma, Non-Small Cell Lung”, “NSCLC”, “Non-Small Cell Lung Cancer”, “ctDNA”, “circulating tumor DNA”, “cell free DNA”, “serum DNA”, “plasma DNA”, “circulating DNA, free DNA”, “free DNA”, “cfDNA”, “prognosis”, “survival”, “prognostic”, “predictive”. Relevant MeSH (Medline) or Emtree (Embase) terms were used where possible. We also hand searched the relevant reference lists to identify new studies. Conference posters and letters that fulfilled the inclusion criteria were also included to capture grey literature. The literature search was confined to English publications.
Two investigators (Fan GW and Zhang K) independently assessed each study for inclusion, and discrepancies were resolved by discussion. Whenever overlapping samples existed (e.g., same authors, overlapping period of study, same protocol ID, overlapping patients), we retained the report with the largest patient population.
Criteria for considering studies for this review
Eligible studies met the following criteria:
dealt with advanced NSCLC (stage IIIB or IV) patients only;
analyzed the correlation between patient survival and EGFR mutations (exon 19 deletions or L858R) and/or KRAS mutations in ctDNA;
had follow-up for overall survival (OS) and/or progression-free survival (PFS); and
provided enough information to obtain HRs directly or indirectly.
Both prospective and retrospective studies were included. Reviews, comments, and case reports were excluded. Studies with less than five patients were also excluded.
Data extraction
Two investigators (Fan GW and Ding JS) independently screened the eligible studies and extracted data using a predefined information sheet that included the following information: first author, publication year, country where the study was conducted, publication type, study design, patients included in analysis, median age, percentage of males, percentage of non-smokers, tumor stage, treatment, detection sample, the time of sampling, ctDNA detection, detection methods, and number of patients with positive ctDNA. Discrepancies were resolved by discussion.
Quality assessment
We evaluated the quality of included studies using the Cochrane Collaboration's tool for assessing risk of bias [32]. Specifically, studies were judged on (1) selection bias: studies that had an explicit statement of inclusion and exclusion criteria were rated as low risk of selection bias; (2) accuracy of exposure assessment, also called measure bias: studies with an explicit statement regarding ctDNA detection methods were rated as low risk; (3) accuracy of other variable assessment: other molecular alterations such as epigenetic alterations and EGFR T790M also influence the prognosis of NSCLC patients; (4) accuracy of outcome assessment; (5) bias due to incomplete follow-up (e.g., median follow-up length, range, and loss-to-follow-up rate were satisfactorily reported); (6) confounding, which included known or commonly discussed confounders in the relationship between ctDNA and survival, such as age, smoking, or other factors that were adjusted.
Measures of treatment effect
The primary outcome was PFS. The secondary outcome was OS. Two investigators (Fan GW, Zhang K) extracted the HRs and their 95% confidence interval (95% CI) to assess the prognostic value of EGFR and KRAS mutations in ctDNA. If HRs for ctDNA were not available, we calculated them indirectly using the methods of Parmar [33]. By convention, HR = 1 indicates a lack of association between ctDNA status and prognosis; HR > 1 indicates a worse survival for patients with ctDNA positive; HR < 1 represents a benefit outcome for the ctDNA-positive group.
Statistical analysis
We used the chi-square test to detect heterogeneity and I2 statistic to measure heterogeneity. A p value > 0.10 and an I2 < 50% indicated a lack of significant heterogeneity; then, the fixed-effects model was used to calculate the pooled HR. Otherwise, the random-effects model was adopted. The effect of ctDNA status on survival was considered statistically significant if the 95% CI for the overall HR estimate did not contain 1.
Sensitivity analyses were conducted for the meta-analysis to check for stability of the overall results. Subgroup analyses were performed using a random-effects model because of the diverse clinical heterogeneity.
Analysis was carried out using Review Manager 5.3 and Stata 12.
Ethics statement
This study was a literature-based study and no ethics approval was needed.
SUPPLEMENTARY MATERIALS FIGURES
Acknowledgments
We thank Dr. Sun Min for his kind help with the data analysis.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no competing interest.
GRANT SUPPORT
This work was supported by the Special Fund for Health-scientific Research in the Public Interest from National Population and family Planning Commission of the P R China (No. 201402018).
REFERENCES
- 1.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. The lancet oncology. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 3.Nygaard AD, Spindler KLG, Pallisgaard N, Andersen RF, Jakobsen A. The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung Cancer. 2013;79:312–317. doi: 10.1016/j.lungcan.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Chen JY, Cheng YN, Han L, Wei F, Yu WW, Zhang XW, Cao S, Yu JP. Predictive value of K-ras and PIK3CA in non-small cell lung cancer patients treated with EGFR-TKIs: a systemic review and meta-analysis. Cancer biology & medicine. 2015;12:126. doi: 10.7497/j.issn.2095-3941.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barlesi F, Mazieres J, Merlio JP, Debieuvre D, Mosser J, Léna H, Ouafik LH, Besse B, Rouquette I, Westeel V. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT) The Lancet. 2016;387:1415–1426. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 6.Camps C, Sirera R, Bremnes R, Blasco A, Sancho E, Bayo P, Safont MJ, Sánchez JJ, Tarón M, Rosell R. Is there a prognostic role of K-ras point mutations in the serum of patients with advanced non-small cell lung cancer? Lung cancer. 2005;50:339–346. doi: 10.1016/j.lungcan.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Huang Q, Li J, Sun Y, Wang R, Cheng X, Chen H. Efficacy of EGFR tyrosine kinase inhibitors in the adjuvant treatment for operable non-small cell lung cancer by a meta-analysis. CHEST Journal. 2016;149:1384–1392. doi: 10.1016/j.chest.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 8.Lee CK, Brown C, Gralla RJ, Hirsh V, Thongprasert S, Tsai CM, Tan EH, Ho JCM, Zaatar A, Sanchez JAO. Impact of EGFR inhibitor in non-small cell lung cancer on progression-free and overall survival: a meta-analysis. Journal of the National Cancer Institute. 2013;105:595–605. doi: 10.1093/jnci/djt072. [DOI] [PubMed] [Google Scholar]
- 9.Kuan FC, Kuo LT, Chen MC, Yang CT, Shi CS, Teng D, Lee KD. Overall survival benefits of first-line EGFR tyrosine kinase inhibitors in EGFR-mutated non-small-cell lung cancers: a systematic review and meta-analysis. British journal of cancer. 2015;113:1519–28. doi: 10.1038/bjc.2015.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA. Circulating mutant DNA to assess tumor dynamics. Nature medicine. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. Journal of Clinical Oncology. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siravegna G, Bardelli A. Blood circulating tumor DNA for non-invasive genotyping of colon cancer patients. Molecular Oncology. 2016;10:475–480. doi: 10.1016/j.molonc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao C, Yuan JQ, Yang ZY, Fu XH, Wu XY, Tang JL. Blood as a Substitute for Tumor Tissue in Detecting EGFR Mutations for Guiding EGFR TKIs Treatment of Nonsmall Cell Lung Cancer: A Systematic Review and Meta-Analysis. Medicine. 2015:94. doi: 10.1097/MD.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jo P, Jung K, Grade M, Conradi LC, Wolff HA, Kitz J, Becker H, Rüschoff J, Hartmann A, Beissbarth T. CpG island methylator phenotype infers a poor disease-free survival in locally advanced rectal cancer. Surgery. 2012;151:564–570. doi: 10.1016/j.surg.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Xu F, Wu J, Xue C, Zhao Y, Jiang W, Lin L, Wu X, Lu Y, Bai H, Xu J. Comparison of different methods for detecting epidermal growth factor receptor mutations in peripheral blood and tumor tissue of non-small cell lung cancer as a predictor of response to gefitinib. Onco Targets Ther. 2012;5:439–447. doi: 10.2147/OTT.S37289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Punnoose EA, Atwal S, Liu W, Raja R, Fine BM, Hughes BG, Hicks RJ, Hampton GM, Amler LC, Pirzkall A. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: Association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clinical Cancer Research. 2012;18:2391–2401. doi: 10.1158/1078-0432.CCR-11-3148. [DOI] [PubMed] [Google Scholar]
- 17.Huang Z, Wang Z, Bai H, Wu M, An T, Zhao J, Yang L, Duan J, Zhuo M, Wang Y. The detection of EGFR mutation status in plasma is reproducible and can dynamically predict the efficacy of EGFR-TKI. Thoracic Cancer. 2012;3:334–340. doi: 10.1111/j.1759-7714.2012.00133.x. [DOI] [PubMed] [Google Scholar]
- 18.Qin L, Zhong W, Zhang L, Ly Li, Wang Mz. Comparison of three methods for detecting epidermal growth factor receptor mutations in plasma DNA samples of Chinese patients with advanced non-small cell lung cancer. Chinese Medical Journal-Beijing. 2011;124:887. [PubMed] [Google Scholar]
- 19.Mack PC, Holland WS, Burich RA, Sangha R, Solis LJ, Li Y, Beckett LA, Lara PN, Davies AM, Gandara DR. EGFR mutations detected in plasma are associated with patient outcomes in erlotinib plus docetaxel-treated non-small cell lung cancer. Journal of Thoracic Oncology. 2009;4:1466–1472. doi: 10.1097/JTO.0b013e3181bbf239. [DOI] [PubMed] [Google Scholar]
- 20.He C, Liu M, Zhou C, Zhang J, Ouyang M, Zhong N, Xu J. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. International Journal of Cancer. 2009;125:2393–2399. doi: 10.1002/ijc.24653. [DOI] [PubMed] [Google Scholar]
- 21.Bai H, Mao L, Zhao J, Yang L, Wang X, Wang J. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. Journal of Clinical Oncology. 2009;27:2653–2659. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- 22.Kimura H, Suminoe M, Kasahara K, Sone T, Araya T, Tamori S, Koizumi F, Nishio K, Miyamoto K, Fujimura M. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA) British journal of cancer. 2007;97:778–784. doi: 10.1038/sj.bjc.6603949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhuo M, Wu M, Zhao J, Song SW, Bai H, Wang S, Yang L, An T, Wang X, Duan J. Epidermal growth factor receptor genotype in plasma DNA and outcome of chemotherapy in the Chinese patients with advanced non-small cell lung cancer. Chinese medical journal. 2011;124:3510–3514. [PubMed] [Google Scholar]
- 24.Camps C, Jantus-Lewintre E, Cabrera A, Blasco A, Sanmartín E, Gallach S, Caballero C, del Pozo N, Rosell R, Guijarro R. The identification of KRAS mutations at codon 12 in plasma DNA is not a prognostic factor in advanced non-small cell lung cancer patients. Lung Cancer. 2011;72:365–369. doi: 10.1016/j.lungcan.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Kim ST, Sung JS, Jo UH, Park KH, Shin SW, Kim YH. Can mutations of EGFR and KRAS in serum be predictive and prognostic markers in patients with advanced non-small cell lung cancer (NSCLC)? Medical Oncology. 2013;30:1–10. doi: 10.1007/s12032-012-0328-3. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez JL, Sarries C, de Castro PL, Roig B, Queralt C, Escuin D, de Aguirre I, Sanchez JM, Manzano JL, Margelí M. Methylation patterns and K-ras mutations in tumor and paired serum of resected non-small-cell lung cancer patients. Cancer letters. 2003;193:207–216. doi: 10.1016/s0304-3835(02)00740-1. [DOI] [PubMed] [Google Scholar]
- 27.Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clinical chemistry. 2015;61:64–71. doi: 10.1373/clinchem.2014.223040. [DOI] [PubMed] [Google Scholar]
- 28.Tursz T, Bernards R. Hurdles on the road to personalized medicine. Molecular oncology. 2015;9:935–939. doi: 10.1016/j.molonc.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nature reviews Clinical oncology. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 30.Zhai Z, Yu X, Yang B, Zhang Y, Zhang L, Li X, Sun H. Colorectal cancer heterogeneity and targeted therapy: Clinical implications, challenges and solutions for treatment resistance. Seminars in Cell & Developmental Biology. 2017;64:107–115. doi: 10.1016/j.semcdb.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 31.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nature Reviews Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 32.Higgins J, Green S. Cochrane handbook for systematic reviews of interventions 4.2. 5 [updated May 2005] The cochrane library. 2005 [Google Scholar]
- 33.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statistics in medicine. 1998;17:2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.