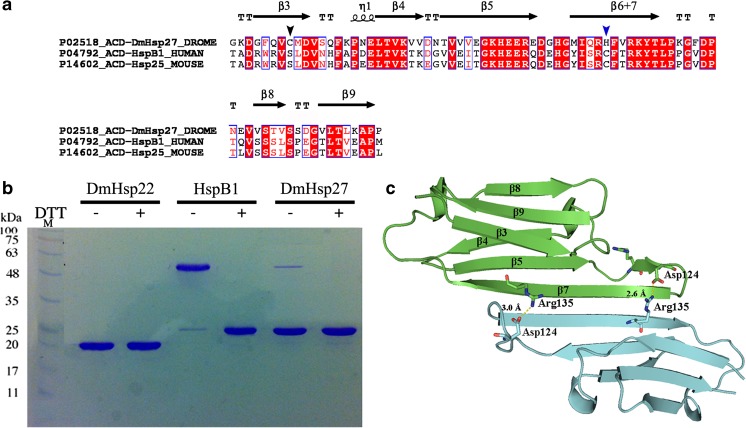
Fig. 1.
Dimeric interface of DmHsp27. a Amino acid sequence alignment of ACD of DmHsp27, human HspB1, and mouse Hsp25. Alignment was generated using multiple sequence comparison by log-expectation (MUSCLE). Secondary structures indicated in the alignment were assigned according to the determined crystal structure of ACD HspB1 (PDB: 4MJH). Black and blue arrowheads indicate cysteine positions. b Non-reducing SDS-PAGE, showing disulfide cross-linking of full-length DmHsp22, DmHsp27, and human HspB1. The proteins were incubated at 20 °C for 21 h with 5 mM DTT (+) or without any reducing agent (−) loaded on non-reducing SDS-PAGE and detected by Coomassie blue staining. c Predicted 3D structure of ACD of DmHsp27 generated with SWISS-MODEL (Biasini et al. 2014) based on defined structure of HspB1 ACD (PDB 4MJH). Visualization of 3D models was realized with PyMOL software. Monomers represented as blue and green. The salt bridge between conserved arginine 135 and aspartic acid 124 is shown