Figure 3.
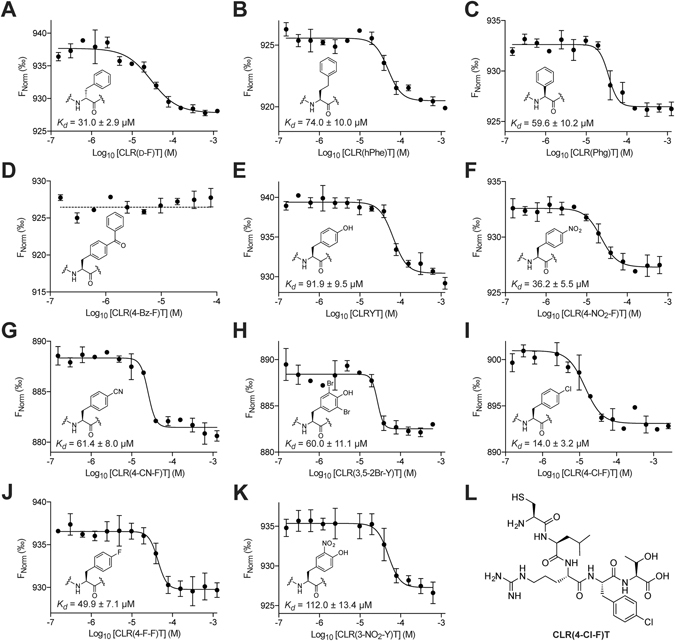
MST analysis of the affinity of CLRFT analogues containing the illustrated non-natural phenylalanine derivatives in place of phenylalanine. (A) The D-phenylalanine analogue binds to CMG238–218 with a K d of 31.0 ± 2.9 µM. (B) The homophenylalanine analogue binds to CMG238–218 with a K d of 74.0 ± 10.0 µM. (C) The phenylglycine analogue binds to CMG238–218 with a K d of 59.6 ± 10.2 µM. (D) The 4-benzoyl-phenylalanine analogue does not bind to CMG238–218. (E) The tyrosine analogue binds to CMG238–218 with a K d of 91.9 ± 9.5 µM. (F) The 4-nitro-phenylalanine analogue binds to CMG238–218 with a K d of 36.2 ± 5.5 µM. (G) The 4-cyano-phenylalanine analogue binds to CMG238–218 with a K d of 61.4 ± 8.0 µM. (H) The 3,5-dibromo-tyrosine analogue binds to CMG238–218 with a K d of 60.0 ± 11.1 µM. (I) The 4-chlorophenylalanine analogue binds to CMG238–218 with a K d of 14.0 ± 3.2 µM. (J) The 4-fluoro-phenylalanine analogue binds to CMG238–218 with a K d of 49.9 ± 7.1 µM. (K) The 3-nitro-tyrosine analogue binds to CMG238–218 with a K d of 112.0 ± 13.4 µM. (L) The structure of the most potent analogue CLR(4-Cl-F)T. All data represented as mean ± SEM, n = 3.
