Abstract
Phased small interfering RNAs (phasiRNAs) are encoded by a novel class of genes known as phasiRNA producing (PHAS) genes. These genes play important regulatory roles by targeting protein coding transcripts in plant species. In this study, 91 regions were identified as potential PHAS loci in tomato, with additional evidence that seven of them can be triggered by five miRNAs. Among the identified loci, 51 were located in genic regions, and the remaining 40 were located in intergenic regions. The transient overexpression of PHAS15 and PHAS26 demonstrated that phasiRNAs predicted by PhaseTank were indeed generated from their respective PHAS loci. Using sRNA-seq data from B. cinerea-infected tomato leaves, we identified 50 B. cinerea-responsive phasiRNAs with increased abundance and five with decreased abundance. Moreover, 164 targets of these differentially expressed phasiRNAs were predicted, and 94 of them were confirmed experimentally using degradome data. Gene ontology analysis of the targets revealed an enrichment of genes with functions related to defense responses and signaling regulation. These results suggest that a large number of endogenous siRNAs, such as phasiRNAs, have not yet been identified in tomato and underscore the urgent need to systematically identify and functionally analyze siRNAs in tomato.
Introduction
Endogenous small RNAs in plants regulate gene expression from DNA modifications to RNA stability at the transcriptional and post-transcriptional levels. These small RNAs include microRNAs (miRNAs); heterochromatic small-interfering RNAs; phased, secondary, small-interfering RNAs (phasiRNAs); and small-interfering RNAs from natural antisense transcripts. In addition to the numerous miRNA studies reported thus far, there has been increasing interest in the identification of loci that generate phasiRNAs. The first of these to be described were the trans-acting small-interfering RNAs (tasiRNAs); thus, tasiRNAs are phasiRNAs that have been shown to target a transcript in trans. Transcripts from these loci are converted into double-stranded RNA (dsRNA) by RNA-dependent RNA polymerase 6 (RDR6)-SGS3 and processed by DCL4, generating siRNAs in a 21-nt phase1–5. These phased siRNAs are known as phasiRNAs, and the loci that produce them were designated as PHAS genes by Zhai et al.6.
Similar to miRNAs, tasiRNAs repress their target transcripts at the post-transcriptional level. As indicated above, the primary transcripts from tasiRNA loci are used to generate dsRNAs by RDR6, and the dsRNAs are cleaved by DCL4 into phased 21-nt segments3–5. Four tasiRNA gene families, TAS1, TAS2, TAS3, and TAS4, have been identified in Arabidopsis thaliana 3, 5, 7. TAS3 is conserved in various plant species8. Aside from these four tasiRNAs generated from non-coding genes, accumulated evidence suggests that several coding genes, such as those encoding PPR, NB-LRR disease resistance proteins, and MYB transcription factors, also generate phased siRNAs5, 6, 9–16.
PhasiRNAs are known to be involved in abiotic17, 18 and biotic stress responses12. In Solanaceae plants, several miRNAs can trigger phasiRNA production from NB-LRR loci in response to biotic stress. Moreover, the tasNB-LRR regulator cascades triggered by miR482 family members are conserved pathways in many plants, particularly in Solanaceae species11, 12. Shivaprasad et al. showed that miR482a can trigger the generation of phasiRNAs from an NBS-LRR gene and enhance the infection of Pseudomonas syringae DC300011. Yang et al. showed that overexpressed miR482e in potato increased plant sensitivity to Verticillium dahliae infection through NBS-LRR targeting and secondary phasiRNA generation19. In a previous study, 90 PHAS loci were predicted using the PhaseTank pipeline on the tomato genome; however, no further analysis for these candidates was presented, such as locus information, chromosome distribution, or experimental evidence20. To identify Botrytis cinerea-responsive phasiRNAs in tomato in this study, we used the PhaseTank pipeline for identification of PHAS loci, with two more sRNA datasets (from mock- and B. cinerea-infected tomato leaves) compared to our previous study20. The candidate genes were also characterized and annotated in terms of overlap with annotated genes, chromosome distribution, and tissue expression. RT-PCR was performed to detect and validate the primary transcripts and to investigate their tissue expression. Two candidates were selected for experimental validation, and the results demonstrated that phasiRNAs predicted by PhaseTank were indeed generated from their respective PHAS loci. Finally, phasiRNAs responsive to B. cinerea were investigated based on sRNA-seq data, and their targets were characterized using degradome data and gene ontology enrichment analysis.
Results
Identification and annotation of PHAS loci and phasiRNAs in tomato
For genome-wide detection of phased sRNAs, PhaseTank21 was employed along with sRNA libraries and tomato genome sequences. A total of 91 sequences were identified as potential PHAS loci in the tomato genome (Tables S1 and S2), with 51 and 40 loci located in annotated genic regions and intergenic regions, respectively. Thirty-one loci were related with disease resistance, including 28R genes; the remaining three were other types of disease resistance genes (Table S2). Among the 40 intergenic PHAS loci, 18 passed the protein-coding-score test and were considered as potential coding RNAs according to the Coding Potential Calculator (CPC) (Table S2). ORF prediction of the remaining 22 intergenic PHAS loci revealed that all of them were less than 100 amino acids in length. According to a previous report, these genes with unknown functions are lincRNA genes21. This phenomenon has also been reported in recent studies on rice22.
Identification of putative phasiRNA triggers
For these 91 PHAS loci, miRNA binding site prediction was performed using the psRNAtarget pipeline (Table S3). Degradome datasets produced from tomato leaves, roots, and fruits were applied as the experimental supports for the identification of potential phasiRNA triggers. Seven PHAS loci (PHAS23, PHAS43, PHAS07, PHAS39, PHAS71, PHAS48, and PHAS51) targeted by five miRNA triggers were subsequently confirmed (Fig. 1).
Figure 1.
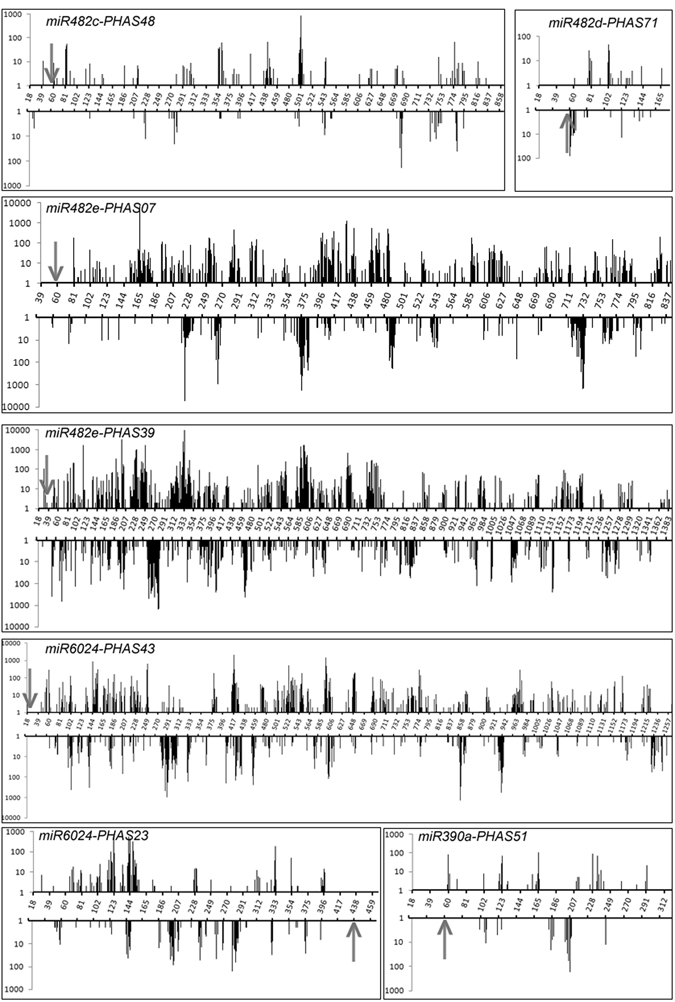
Read abundance distribution of seven PHAS loci in the sRNA library. The arrows indicate the miRNA cleavage sites. The X-axis represents a double-stranded PHAS locus. Abscissa indicates the phasiRNA position mapped within the PHAS locus. Ordinate indicates the read abundance of the mapped sRNAs.
Transcript activity analysis of 91 PHAS loci
Nearly half of the 91 putative PHAS genes were located in intergenic regions (40 loci). Their transcriptional activity was therefore investigated to assess their biogenesis and biological functionusing transcriptome data from seedlings, flowers, roots, and fruits and Tophat23 and Cufflinks24 analysis. Thirty-five of the 91 PHAS loci were transcribed in these tissues; the remaining 56 candidates were not detected (Table S2).
RT-PCR was also used to detect the transcript levels of the 91 PHAS loci. Thirty-seven PHAS genes were amplified in a mixed tissue sample of tomato roots, stems, and leaves (Table 1 and Figure S1). No amplicons were detected for the remaining 54 PHAS candidates. The 37 amplified PHAS genes were sequenced for validation and submitted to GenBank (accession numbers KU555888–KU555924) (Table 1). Except for Sly-PHAS08, Sly-PHAS37, and Sly-PHAS38, sequence analysis revealed that all amplicons had >99% alignment, confirming the transcriptional activities of 34 PHAS gene loci. Interestingly, only 15 PHAS candidates could be detected using both transcriptome data and RT-PCR. Nineteen candidates, which were not detected from the transcriptome data, were amplified in the mixed samples, suggesting tissue-specific transcription in tomato stems. The remaining 57 PHAS candidates were not detected, and their transcript levels could not be validated by RT-PCR. The failure to detect these putative PHAS genes may be attributable to expression at extremely low levels or in a highly tissue-specific manner. Moreover, this fraction of phasiRNAs may not be amenable to confirmation through these means.
Table 1.
Transcriptional confirmation and tissue expression of 91 putative PHAS genes based on RT-PCR.
| PHAS_ID | Accession No. | Root | Stem | Leaf | PHAS_ID | Accession No. | Root | Stem | Leaf |
|---|---|---|---|---|---|---|---|---|---|
| PHAS02 | KU555888 | √ | √ | √ | PHAS21 | KU555907 | √ | √ | √ |
| PHAS03 | KU555889 | √ | √ | √ | PHAS22 | KU555908 | √ | √ | √ |
| PHAS04 | KU555890 | √ | √ | √ | PHAS23 | KU555909 | √ | √ | √ |
| PHAS05 | KU555891 | No | √ | √ | PHAS24 | KU555910 | √ | √ | √ |
| PHAS06 | KU555892 | √ | √ | √ | PHAS25 | KU555911 | √ | √ | √ |
| PHAS07 | KU555893 | √ | √ | √ | PHAS26 | KU555912 | √ | √ | √ |
| PHAS08 | KU555894 | No | √ | No | PHAS27 | KU555913 | √ | √ | √ |
| PHAS09 | KU555895 | √ | √ | √ | PHAS28 | KU555914 | √ | √ | No |
| PHAS10 | KU555896 | √ | √ | √ | PHAS29 | KU555915 | √ | √ | √ |
| PHAS11 | KU555897 | √ | √ | √ | PHAS30 | KU555916 | √ | √ | √ |
| PHAS12 | KU555898 | No | √ | No | PHAS31 | KU555917 | √ | √ | √ |
| PHAS13 | KU555899 | √ | √ | √ | PHAS32 | KU555918 | √ | √ | √ |
| PHAS14 | KU555900 | √ | √ | √ | PHAS33 | KU555919 | √ | √ | √ |
| PHAS15 | KU555901 | √ | √ | √ | PHAS34 | KU555920 | √ | √ | √ |
| PHAS16 | KU555902 | No | No | √ | PHAS35 | KU555921 | No | √ | √ |
| PHAS17 | KU555903 | No | √ | √ | PHAS36 | KU555922 | √ | √ | √ |
| PHAS18 | KU555904 | √ | √ | √ | PHAS37 | KU555923 | √ | √ | √ |
| PHAS19 | KU555905 | √ | √ | √ | PHAS38 | KU555924 | √ | No | √ |
| PHAS20 | KU555906 | √ | √ | √ |
Experimental validation of two PHAS loci and their phasiRNAs
If the analysis above is correct, the transcription of the identified PHAS genes should be possible in tomato along with the production of their phasiRNAs. To test this, two PHAS loci (PHAS15 and PHAS26) and their phasiRNAs (siR15-2 and siR15-6 from PHAS15 and siR26-2 and siR26-4 from PHAS26) were selected for validation through transient expression in tomato leaves and detection of the phasiRNAs. PHAS15 is located at the Solyc06g054600.2 locus, which encodes zinc finger CCCH domain-containing protein 58, while PHAS26 is intergenic. A RACE kit was used to amplify the respective 5’ and 3’ sequences, and the results revealed that both PHAS15 and PHAS26 were transcribed from the negative strand. This means that PHAS15 was not from the genic region of Solyc06g054600.2 (Table S2); instead, it was encoded by the antisense strand of Solyc06g054600.2 (Fig. 2). According to the 5’ and 3’ sequences of PHAS15 and PHAS26, both full-length cDNAs were also amplified from tomato leaves and sequenced (Fig. 2). The sequence analysis showed that PHAS15 is 1193 bp in length and located on chromosome 6 from 37325436 to 37326628, partially overlapping the antisense strand of Solyc06g054600.2. PHAS26 is 1020 bp in length and located in an intergenic region of chromosome 11 from 18739580 to 18740598 (Figure S2).
Figure 2.
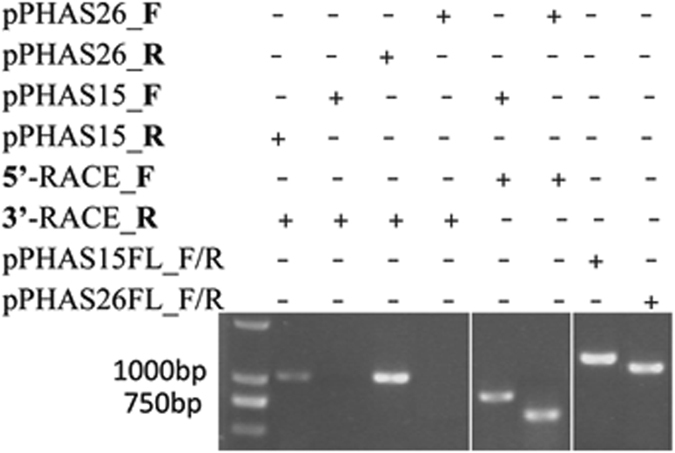
RACE analysis for PHAS15 and PHAS26.
Next, overexpression vectors of PHAS15 and PHAS26 were constructed by fusing PHAS15 and PHAS26 with the CaMV35S promoter (Fig. 3A). Agrobacterium tumefaciens LBA4404 cells containing either the PHAS15 or PHAS26 overexpression vectors were injected into tomato leaves on one side of the midvein. LBA4404 cells with blank pEarleyGate100 (PEG100) vector were also cultured and injected into tomato leaves on one side of the midvein as the control. After 24 h, RT-qPCR analysis was performed. The results showed that PHAS15, PHAS26, and the four phasiRNAs were transiently overexpressed in the leaves transformed with the PHAS fusion vectors, demonstrating that the identified phasiRNAs are indeed generated from the respective PHAS loci (Fig. 3B and C). These results confirmed the PHAS identification from the PhaseTank pipeline.
Figure 3.
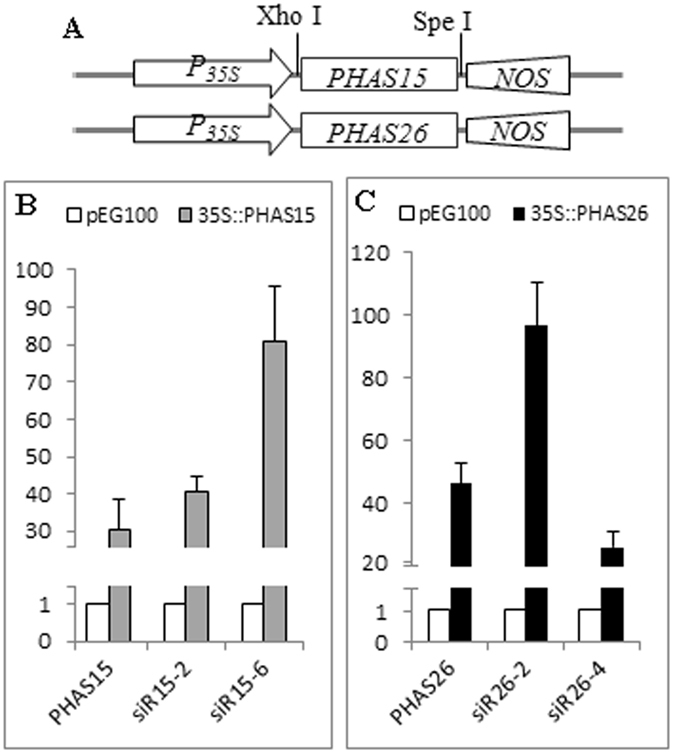
Transient overexpression. (A) Construction of the PHAS overexpression vectors. (B) The expression levels of PHAS15 and the corresponding phasiRNAs were detected in p35S::PHAS15-transformed leaves and control leaves. (C) The expression levels of PHAS26 and the corresponding phasiRNAs were detected in p35S::PHAS26-transformed leaves and control leaves. Ubi3 was used as the internal control. Error bars indicate SD from three biological repeats; the normalized levels of the control were set arbitrarily to 1.
Identification of B. cinerea-responsive phasiRNAs and experimental evidence
As indicated above, 31 of the identified PHAS loci mapped to the coding regions of genes encoding disease resistance-related proteins, suggesting that phasiRNA could be responsive to disease infection. To investigate phasiRNAs responsive to B. cinerea in tomato, reads from two small RNA libraries of tomato leaves inoculated with B. cinerea (infected and control) were analyzed. A total of 245 phasiRNAs with relatively high expression were subjected to further analysis. Among these 245 phasiRNAs, 50 from 27 loci hand increased abundance and five phasiRNAs from five loci had decreased abundance in B. cinerea-infected leaves (Table S4).
To validate these 55 B. cinerea-responsive phasiRNAs, sRNA-seq data from B. cinerea-inoculated tomato leaves at three time points (0 h, 24 h, and 72 h) were used25. Twenty-nine phasiRNAs were confirmed, including 27 phasiRNAs with increased abundance and two with decreased abundance, and the expression patterns were consistent with our sRNA-seq data. Another eight phasiRNAs also had altered expression in B. cinerea-infected leaves, but an opposite change in expression patterns was observed. Among the remaining 18 phasiRNAs, no expression changes were found for ten phasiRNAs, and expression analysis could not be performed for the remaining eight phasiRNAs (siR31-4, siR60-1, siR38-14, siR16-5, siR40-13, siR23-3, siR55-1, and siR46-1) due to their very low read counts (<10 TP10M) (Fig. 4A and Table S5). sRNA-seq data from B. cinerea-infected tomato fruits at the same time points were also used. Among the 29 confirmed phasiRNAs, 23 (22 with increased abundance and one with decreased abundance) were further validated with consistent expression patterns in fruit (Fig. 4B and Table S5) and will thus be analyzed in subsequent works.
Figure 4.
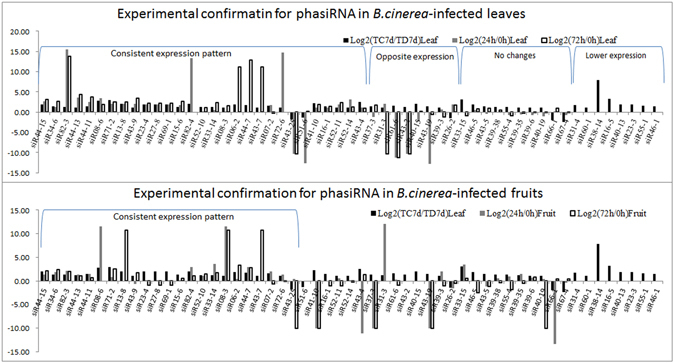
Experimental confirmation for B.cinerea-responsive phasiRNAs in tomato leaves (A) and fruits (B).
From the analysis of phasiRNAs responsive to B. cinerea infection, five phasiRNAs triggered by four conserved miRNAs (miR6024, miR482d, miR482e, and miR390a) were also found. SiR51-6 from miR390a-PHAS51, which was highly homologous to TAS3, had decreased abundance in B. cinerea-infected leaves, but no expression signal was detected in tomato fruits. Otherwise, the remaining four phasiRNAs triggered by conserved miRNAs (siR07-2 triggered by miR482e, siR71-2 triggered by miR482d, and both siR23-4 and siR43-7 triggered by miR6024) had increased abundance in both B. cinerea-infected leaves and B. cinerea-infected fruits (Fig. 4).
Target prediction of phasiRNAs and degradome evidence
To identify the downstream regulatory pathway of the phasiRNAs, their targets were searched by identifying complementary regions from all tomato transcripts. Using psRNAtarget with default parameters, 164 putative targets were found (Table S6) and further validated with degradome data. The analysis showed that the cleavage start positions of 94 targets overlapped with degradome fragments, and these findings were considered as experimental validation (Table S6). GO enrichment analysis indicated that all of the targets are binding proteins related to defense responses or signaling regulation (Fig. 5). Among them, 11 targets were also the site of a PHAS gene locus.
Figure 5.
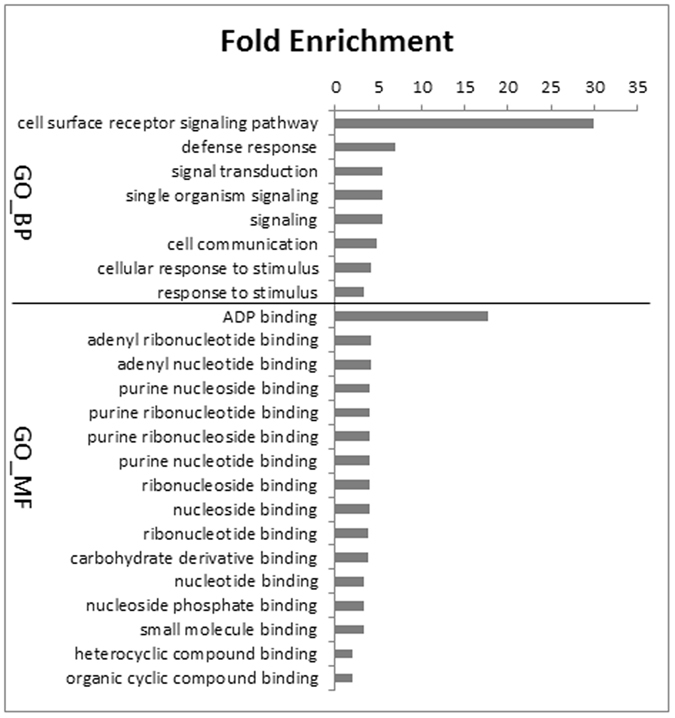
GO functional enrichment analysis of the target genes of B. cinerea-responsive phasiRNAs.
Discussion
PhasiRNAs have been shown to have important roles in developmental regulation and stress responses. Genome-wide identification of phasiRNAs in a given organism is an important tool that can be used to investigate gene regulation involving small RNAs and is as important as mining genes that code for proteins. The establishment of a comprehensive list of phasiRNAs from any organism will be instrumental not only for gene regulation studies but also for genome organization, phylogenetic comparison, comparative development, and other evolutionary analyses. In this study, the PhaseTank pipeline identified 912 phasiRNAs from 91 PHAS loci in the tomato genome. However, only seven PHAS loci (PHAS23, PHAS43, PHAS07, PHAS39, PHAS71, PHAS48, and PHAS51) were identified as the targets of five miRNA triggers and subsequently confirmed based on degradome data. Trigger miRNAs were not found for the remaining 84 PHAS loci, and possible explanations are as follows. 1) The tomato miRNAs deposited into the miRBase database is not complete, and a large number of miRNAs may still be found. Thus far, 713 and 427 miRNAs have been reported in rice and Arabidopsis, respectively. In potato, which is in the same family as tomato, 343 miRNAs have been reported. In contrast, only 110 miRNAs have been deposited into miRBase (version 21.0) for tomato26. Therefore, incomplete information about tomato miRNAs may hinder the identification of trigger miRNAs. 2) Detection failure may also reflect the possibility that many of these PHAS loci are triggered by siRNAs rather than miRNAs.
As first described in Arabidopsis5, phasiRNAs are generated from both protein-coding regions5, 6, 15, 16 and long none-coding regions27–29 in many plant genomes. In this study, 51 coding and 40 non-coding regions were identified as the sites of PHAS loci in tomato. Among the coding regions identified as PHAS loci, NB-LRR was predominantly represented. NB-LRR regulation by secondary siRNAs has been reported to exist widely in Solanaceae species6, 11, 12. Most recently, an examination of numerous NB-LRRs in a wide variety of plant species demonstrated significant levels of secondary siRNAs in Norway spruce (Picea abies, a gymnosperm), Amborella (a basal angiosperm), cotton (Gossypium hirsutum), poplar (Populus spp.), grapevine (Vitis vinifera), apple, and peach, indicating broad conservation and an ancient origin for the role of phasiRNAs in the regulation of NB-LRRs13.
PhasiRNAs have also recently been shown to be involved in biotic stress responses12. In this study, B. cinerea-responsive phasiRNAs, including 27 with increased abundance and two with decreased abundance, were identified and validated in B. cinerea-infected tomato leaves. Moreover, 22 of the 27 phasiRNAs with increased abundance and one of the two phasiRNAs with decreased abundance also had the same expression patterns in B. cinerea-infected tomato fruits. Three miRNA-PHAS regulation cascades (miR482d-PHAS, miR482e-tasLRR, and miR6024-tasLRR) were also identified and confirmed in response to B. cinerea infection. The miR482 family is well-known as a trigger miRNA involved in phasiRNA biogenesis11, 19, 30. In addition, miR6024 may also trigger the production of phasiRNAs at NB-LRR loci in Solanaceae plants12. With the focus on responsiveness to B. cinerea in mind, many phasiRNAs were predicted to target R genes such as NBS-LRR or RLP genes, which are important for pathogen resistance in higher plants. The NBS-LRR gene family is a huge gene family with dozens to hundreds of members per plant genome31, 32. While maintaining a large number of NBS-LRR genes may be beneficial for plant defense against different pathogens, maintaining their expression under a pathogen-absent environment may incur a fitness cost33. Therefore, phasiRNA-targeted R genes may play important roles in biotic stress responses. Putative targets were also found for these B. cinerea-responsive phasiRNAs, including a NAC transcription factor; MADS-box transcription factor; zinc finger CCCH domain-containing protein; and a series of plant signal transduction factors, such as an auxin response factor, serine/threonine protein kinase, and F-box protein. Because the identified phasiRNAs clearly responded to B. cinerea infection in tomato, their validity warrants further investigation, and confirming their differential expression will provide clues about their biological functions.
Finally, to gain insight into the biological effect of the identified B. cinerea-responsive phasiRNAs, PHAS-overexpressing leaves confirmed by qRT-PCR (Fig. 3B and C) were also inoculated with B. cinerea agar discs. After 48 h, the results showed that the phasiRNAs led to larger pathogenic spots in the leaves overexpressing PHAS15 or PHAS26 compared to control leaves (Fig. 6). Although the transient expression results need to be validated by other experiments, the findings suggest that these B. cinerea-responsive phasiRNAs might be involved in the regulation of B. cinerea infection in tomato.
Figure 6.

Biological impact of PHAS15 and PHAS26. Agrobacterium tumefaciens LBA4404 cells containing the overexpression vectors were injected into tomato leaves on one side of the midvein. LBA4404 cells with blank pEG100 vector were also cultured and injected into tomato leaves on one side of the midvein as the control. After 24 h, the opposite sides of the injected leaves were used for inoculation with B. cinerea agar discs (4 mm in diameter) for 48 h. The yellow arrows indicate the inoculation sites of the B. cinerea agar discs.
Materials and Methods
Data retrieval
Three small RNA libraries (GSM803579, GSM803580, and GSM803581) produced from tomato leaves, flowers, and fruit of S. lycopersicum were retrieved from the NCBI Gene Expression Omnibus database under the accession number GSE28755 from Chávez Montes et al.34. Their study examined the conservation and divergence of microRNAs from 99 different tissues from three algal species and 31 representative vascular plant species. Two additional small RNAs libraries (SRR1482408 and SRR1463412) came from seven day post-inoculation leaves of B. cinerea-inoculated and mock-inoculated plants; these data were produced by Jin et al.35 to identify B. cinerea-responsive miRNAs in tomato and retrieved from the NCBI Sequence Read Archive (SRA) database.
Three degradome datasets (GSM553688, GSM553689, and GSM5536903) produced by Lopez-Gomollon et al.36 from tomato leaves, roots, and fruits were retrieved from the NCBI Gene Expression Omnibus database under the accession number GSE22236. In addition, four paired-end mRNA sequencing libraries (SRX1227045, SRX1227046, ERX1604048, and SRX1227066) produced from the seedlings, flowers, roots, and fruits were downloaded from the NCBI SRA database.
Identification of phasiRNA candidate genes
To detect phased sRNAs in tomato, PhaseTank21 was used with default parameters on five sRNA libraries (GSM803579, GSM803580, GSM803581, SRR1482408, and SRR1463412) downloaded from the NCBI SRA database and tomato genome sequences (Version SL2.50) downloaded from the ftp site (ftp://ftp.solgenomics.net/genomes/Solanum_lycopersicum/annotation/).
Transcription activity analysis of 91 PHAS loci
Four paired-end mRNA sequencing libraries (SRX1227045, SRX1227046, ERX1604048, and SRX1227066) produced from tomato leaves, flowers, roots, and fruits were downloaded from the NCBI SRA database. These RNA-seq data were used to analyze the transcription of 91 PHAS loci through Tophat23 and Cufflinks24. Low quality reads were removed with fastq_quality_filter using the options –q 20 and –p 80 (http://hannonlab.cshl.edu/fastx_toolkit/). Filtered reads from each sample were aligned to the tomato reference genome (SL2.50) using Tophat23. The transcriptome of each sample was assembled separately using Cufflinks, and Cuffmerge was used to merge the assemblies produced by Cufflinks for all samples24. If the genomic coordinate of a given PHAS locus overlapped with a merged assembly locus, the transcription of the PHAS locus was considered confirmed.
Protein-coding-score test and open reading frame assay
The Coding Potential Calculator37, which detects the quality, completeness, and sequence similarity of an ORF to proteins in current protein databases, was used to predict putative protein-encoding transcripts of PHAS loci with default parameters. Only loci that did not pass the protein-coding-score test were further analyzed for ORF prediction. A Perl script was written to investigate the intergenic PHAS loci that encode ORFs of 100 or fewer amino acids through six-frame translation.
Target prediction and validation of miRNAs and phasiRNAs
In this report, the targets of miRNAs and phasiRNAs were initially predicted using the online tool psRNATarget38. Next, the cleavage start position was validated by mapping the target sequences with three degradome datasets (GSM553688, GSM553689, and GSM5536903) produced from tomato leaves, roots, and fruits. If any RNA degradome fragment was complementary to the target PHAS of a miRNA and the 5’-end of the degradome fragment overlapped the PHAS cleavage start position, the start position of miRNA-guided PHAS cleavage was considered validated6, 39. Validation of the phasiRNA cleavage site was also performed using the RNA degradome. If any RNA degradome fragment was complementary to the target transcript of a phasiRNA and the 5’-end of the degradome fragment overlapped with the 8–12 positions of the phasiRNA, the cleavage site was considered confirmed.
GO enrichment analyses
GO enrichment analysis was applied to target transcripts using an online tool of the PANTHER Classification System40. GO terms with P < 0.05 were considered significantly enriched.
RNA extraction and RT-PCR analysis
Total RNAs were extracted using TRNzol-A+ Reagent (TIANGEN, Beijing, China), followed by RNase-free DNase treatment (Takara, Dalian, China). The concentrations were quantified using a NanoDrop ND-1000 spectrophotometer.
Reverse transcription was performed using the PrimeScript RT® reagent kit (Perfect Real Time) (TaKaRa, Dalian, China). All PHAS and target genes were subjected to RT-PCR validation and quantitation using specific primers (Table S7), which were designed using Primer Premier 5. The reaction conditions were as follows: 95 °C for 3 min; 30 cycles of 95 °C for 30 s, 49–62 °C for 30 s, and 72 °C for 40 s; and final elongation at 72 °C for 10 min.
Reverse transcription was also performed using the One Step PrimeScript miRNA cDNA Synthesis Kit (TaKaRa, Dalian, China) according to the manufacturer’s protocol for phasiRNA quantitation. All phasiRNAs were subjected to qRT-PCR validation using specific primers (Table S1), which were designed using Primer Premier 5. The reaction conditions were as follows: 94 °C for 4 min; 30 cycles of 94 °C for 30 s, 60 °C for 40 s, and 72 °C for 40 s; and final elongation at 72 °C for 5 min.
Construction of the expression vectors
The full-length cDNAs of PHAS genes were amplified and cloned into pGEM-T. After sequencing validation and digestion, the PHAS sequence was introduced into the pEG100 vector using the Xho I and Spe I restriction sites. The resulting construct contained the PHAS gene driven by the cauliflower mosaic virus 35S (CaMV35S) promoter and terminated by nos (Fig. 3A). The construct was introduced into A. tumefaciens LBA4404 for transient expression in tomato leaves.
Transient overexpression
A. tumefaciens LBA4404 cells transformed with recombinant pEG100-PHAS plasmid was cultured at 28 °C in LB medium supplemented with kanamycin (50 μg/ml) and rifampicin (20 μg/ml). When turbidity at 600 nm reached 1.0, the cells were collected by centrifugation at 5,000 rpm for 5 min and washed twice in resuspension buffer containing 10 mmol MES (pH 5.6), 10 mmol MgCl2, and 200 μmol acetosyringone. The cell pellets were resuspended in resuspension buffer at an OD600 of 0.5 then injected into the tomato leaves. LBA4404 cells transformed with the empty pEG100 vector were also cultured and used in the same manner to serve as the control.
For the transient overexpression of PHAS, tomatoes (Solanum lycopersicum cv. micro-Tom) were used as the host plant. They were grown in a greenhouse with a 16-h day/8-h night cycle at 22–28 °C. At the age of 6 weeks, LBA4404 cells containing the overexpression vectors were injected into tomato leaves on one side of the midvein. LBA4404 cells with blank pEG100 vector were also cultured and injected into tomato leaves on one side of the midvein as the control. After 24 h, the other sides of the injected leaves were harvested in three biological replicates. The samples were frozen in liquid nitrogen and stored at −70 °C for the transcript expression analysis.
Quantitative real-time PCR analysis
Expression profiles of the PHAS genes and phasiRNAs were assayed by quantitative reverse transcription PCR (qRT-PCR). Total RNA was treated with RNase-free DNase I (TaKaRa, Dalian, China) to remove genomic DNA. For the PHAS genes, first-strand cDNA synthesis was performed using SuperScript II Reverse Transcriptase (Invitrogen, USA). For the phasiRNAs, the reverse transcription reaction was performed using the One Step PrimeScript miRNA cDNA Synthesis Kit (TaKaRa, Dalian, China) according to the manufacturer’s protocol41. All of the oligos used in this study are listed in Supplemental Table S7.
SYBR Green PCR was performed as per the manufacturer’s instructions (Takara, Japan). Briefly, 2 μl of cDNA template was added to 12.5 μl of 2 × SYBR Green PCR master mix (Takara), 1 μM concentration of each primer, and ddH2O to a final volume of 25 μl. The reactions were amplified for 10 s at 95 °C, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. All reactions were performed in triplicate, and the controls (no template and no RT) were included for each gene. The threshold cycle (CT) values were automatically determined by the instrument, and the fold change was calculated using the following equation: 2−ΔΔCt, where ΔΔCT = (CT,target − CT,inner)Infection − (CT,target − CT,inner)Mock 42.
Identification of B. cinerea-responsive phasiRNAs
The deep sequencing data for small RNAs in 7 days post-inoculation leaves of B. cinerea-infected (TD7d) and control (TC7d) plants were used (accession numbers SRR1482408 and SRR1463412). The frequency of phasiRNAs was normalized as ‘reads per million’ (RPM = mapped reads/total reads × 1,000,000). The fold_change between the TD7d and TC7d libraries was calculated using the following equation: fold_change = log2 (RPMTD7d/RPMTC7d). PhasiRNAs with fold-changes >1 or <−1 and p-values ≤ 0.001 were considered to have increased abundance or decreased abundance in response to B. cinerea stress, respectively. The p-value was calculated according to previously established methods43.
Electronic supplementary material
Acknowledgements
This work was supported by the Natural Science Foundation of China (grant no. 31372075), the Natural Science Foundation of Zhejiang Province (grant no. LQ15C140021), the Doctoral Scientific Research Foundation of Zhejiang Sci-Tech University (grant nos. 13042160-Y and 13042161-Y), and the 521 Talent Foundation of Zhejiang Sci-Tech University.
Author Contributions
F.W. and W.J. designed and wrote the manuscript. Y.C., X.Z. and X.T. performed experiments. W.J. performed bioinformatics analysis. F.W. collected samples and prepared figures. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02233-x
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen X. Small RNAs in development-insights from plants. Current opinion in genetics & development. 2012;22:361–367. doi: 10.1016/j.gde.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axtell MJ. Classification and comparison of small RNAs from plants. Annual review of plant biology. 2013;64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- 3.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howell MD, et al. Genome-Wide Analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 Pathway in Arabidopsis Reveals Dependency on miRNA- and tasiRNA-Directed Targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhai J, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes & Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 9.Xia R, et al. MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. The Plant Cell. 2013;25:1555–1572. doi: 10.1105/tpc.113.110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H-M, Li Y-H, Wu S-H. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007;104:3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shivaprasad PV, et al. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. The Plant Cell Online. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F, et al. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Källman T, Chen J, Gyllenstrand N, Lagercrantz U. A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. Plant Physiol. 2013;162:741–754. doi: 10.1104/pp.113.214643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu QH, et al. miR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PloS ONE. 2013;8:e84390. doi: 10.1371/journal.pone.0084390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia R, Zhu H, An Y-Q, Beers EP, Liu Z. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biology. 2012;13:R47. doi: 10.1186/gb-2012-13-6-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H, et al. Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC plant biology. 2012;12:149. doi: 10.1186/1471-2229-12-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh LC, et al. Uncovering Small RNA-Mediated Responses to Phosphate Deficiency in Arabidopsis by Deep Sequencing. Plant Physiol. 2009;151:2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo QJ, Mittal A, Jia F, Rock CD. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 2012;80:117–129. doi: 10.1007/s11103-011-9778-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, et al. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 2015;57:1078–1088. doi: 10.1111/jipb.12348. [DOI] [PubMed] [Google Scholar]
- 20.Guo Q, Qu X, Jin W. PhaseTank: genome-wide computational identification of phasiRNAs and their regulatory cascades. Bioinformatics. 2015;31:284–286. doi: 10.1093/bioinformatics/btu628. [DOI] [PubMed] [Google Scholar]
- 21.Li L, et al. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 2014;15:R40. doi: 10.1186/gb-2014-15-2-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komiya R, et al. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 2014;78:385–397. doi: 10.1111/tpj.12483. [DOI] [PubMed] [Google Scholar]
- 23.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiberg A, et al. Fungal Small RNAs Suppress Plant Immunity by Hijacking Host RNA Interference Pathways. Science. 2013;342:118. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Grocock RJ, van, Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding J, et al. RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol. Plant. 2012;5:1210–1216. doi: 10.1093/mp/sss095. [DOI] [PubMed] [Google Scholar]
- 28.Ding J, et al. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA. 2012;109:2654–2659. doi: 10.1073/pnas.1121374109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou H, et al. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 2012;22:649–660. doi: 10.1038/cr.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F, Orban R, Baker B. SoMART: a web server for plant miRNA, tasiRNA and target gene analysis. The plant Journal. 2012;70:891–901. doi: 10.1111/j.1365-313X.2012.04922.x. [DOI] [PubMed] [Google Scholar]
- 31.Shao ZQ, et al. Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: understanding gained from and beyond the legume family. Plant Physiol. 2014;166:217–234. doi: 10.1104/pp.114.243626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YM, et al. Uncovering the dynamic evolution of nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes in Brassicaceae. J. Integr. Plant Biol. 2016;58:165–177. doi: 10.1111/jipb.12365. [DOI] [PubMed] [Google Scholar]
- 33.Tian D, et al. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- 34.Chávez Montes RA, et al. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun. 2014;5:3722. doi: 10.1038/ncomms4722. [DOI] [PubMed] [Google Scholar]
- 35.Jin W, Wu F. Characterization of miRNAs associated with Botrytis cinerea infection of tomato leaves. BMC Plant Biol. 2015;15:1. doi: 10.1186/s12870-014-0410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Gomollon S, et al. Diverse correlation patterns between microRNAs and their targets during tomato fruit development indicates different modes of microRNA actions. Planta. 2012;236:1875–1887. doi: 10.1007/s00425-012-1734-7. [DOI] [PubMed] [Google Scholar]
- 37.Kong L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;36:W345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang CQ, Li GP, Wang J, Fang JG. Identification of trans-acting siRNAs and their regulatory cascades in grapevine. Bioinformatics. 2012;28:2561–2568. doi: 10.1093/bioinformatics/bts500. [DOI] [PubMed] [Google Scholar]
- 40.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang TZ, Chen L, Zhao MG, Tian QY, Zhang WH. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics. 2011;12:367. doi: 10.1186/1471-2164-12-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(t)Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Man MZ, Wang X, Wang Y. POWER_SAGE: comparing statistical tests for SAGE experiments. Bioinformatics. 2000;16:953–959. doi: 10.1093/bioinformatics/16.11.953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


