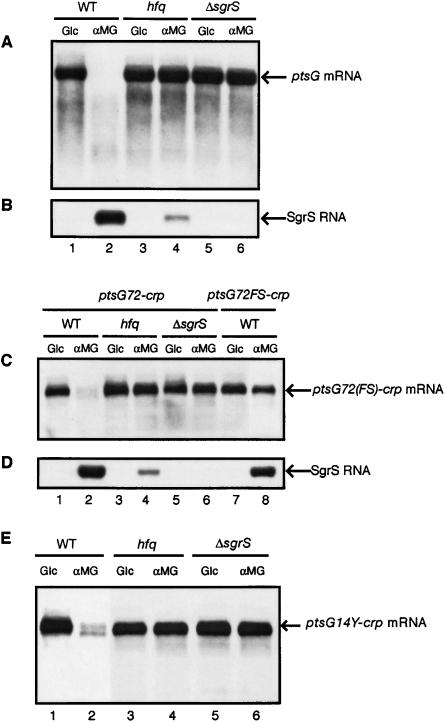
Figure 8.
Effects of hfq or sgrS mutation on the degradation of ptsG and ptsG-crp mRNAs. (A,B) AS16, AS21 (ΔptsG hfq::cat), and AS23 (ΔptsG ΔsgrS) harboring pTH111 were grown to A600 = 0.5. Glc, or αMG was added at a final concentration of 1%, and incubation was continued for 20 min. Total RNAs were prepared and 3 μg of RNA samples was subjected to Northern blot analysis using ptsG or sgrS probes. (C,D) IT1568, TM122 (hfq::cat), and TM542 (ΔsgrS) harboring pMM12, and IT1568 harboring pMM12FS were grown to A600 = 0.5. Glc, or αMG was added at a final concentration of 1%, and incubation was continued for 20 min. Total RNAs were prepared and 3 μg of RNA samples was subjected to Northern blot analysis using crp or sgrS probes. (E) IT1568, TM122, and TM542 harboring pMM10Y were grown to A600 = 0.5. Glc or αMG was added at a final concentration of 1%, and incubation was continued for 20 min. Total RNAs were prepared and 3 μg of RNA samples was subjected to Northern blot analysis using the crp probe.