Figure 4.
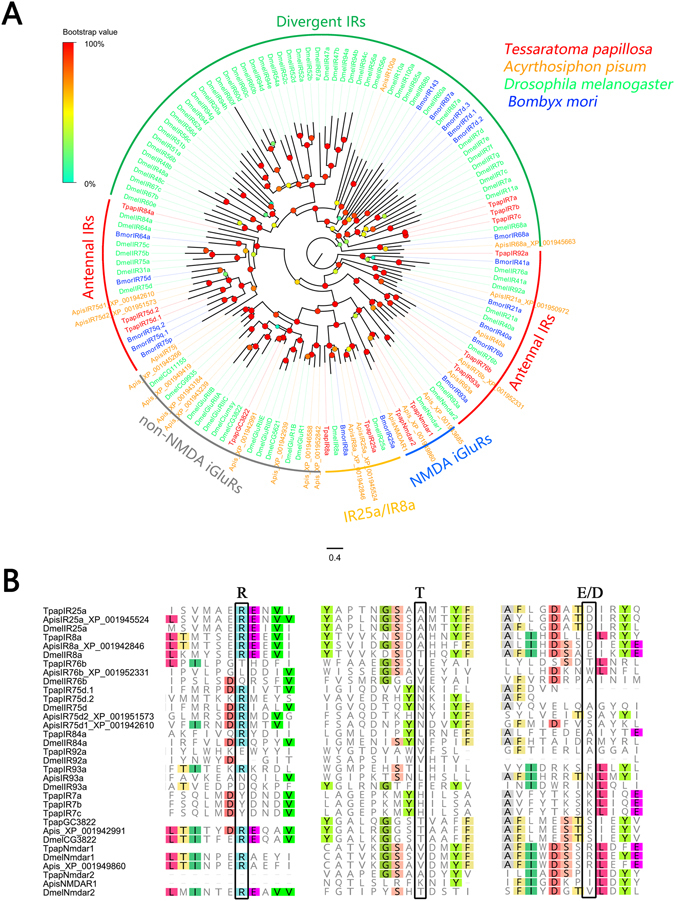
(A) Phylogenetic analysis of putative T. papillosa iGluRs/IRs in with other insect iGluRs/IRs. The dendrogram was generated by FastTree2 (JTT substitution model). Species abbreviations: Tpap, Tessaratoma papillosa; Apis, Acyrthosiphon pisum; Bmor, Bombyx mori; Dmel, Drosophila melanogaster. Amino acid sequences used to construct the tree are shown in Supplementary Table S2. Branch support (circles at the branch nodes) was estimated using an approximate likelihood ratio test based on the scale indicated at the top left. Bars indicate branch lengths in proportion to amino acid substitutions per site. (B) Excerpts from the amino acids alignment showing the predicted iGluRs/IRs binding domains.
