Figure 6.
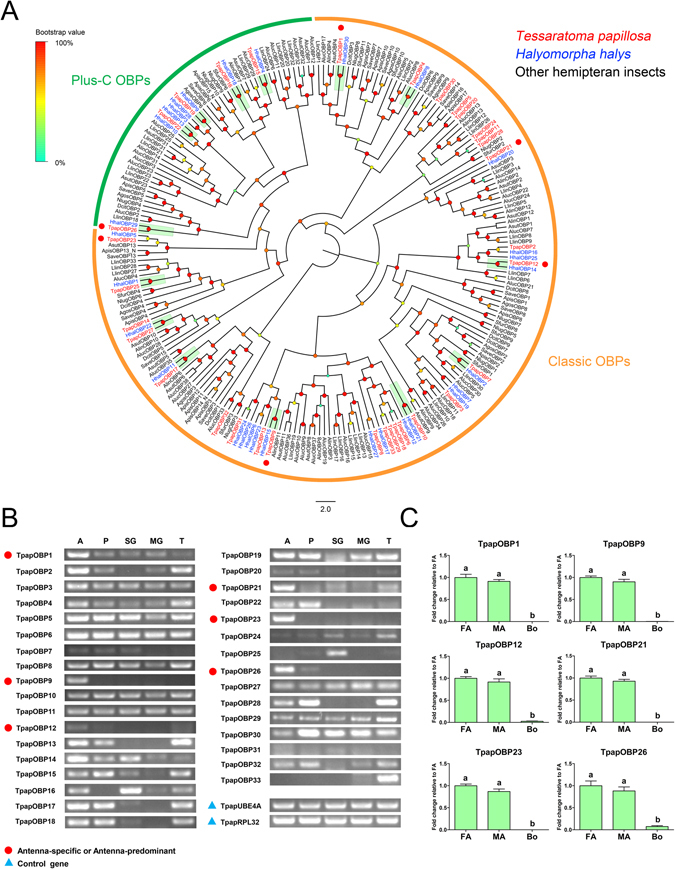
(A) Phylogenetic analysis of putative T. papillosa OBPs with other hemipteran OBPs. Plus-C OBPs from T. papillosa and other putative hemipterans form a clade labeled in dark green. Labeled in orange are classic OBPs from T. papillosa and other putative hemipterans. Species abbreviations: Tpap, Tessaratoma papillosa; Hhal, Halyomorpha halys; Llin, Lygus lineolaris; Aluc, Apolygus lucorum. Alin, Adelphocoris lineolatus, Asut, Adelphocoris suturalis, Save, Sitobion avenae, Apis, Acyrthosiphon pisum, Agos, Aphis gossypii, Nlug, Nilaparvata lugens, Sfur, Sogatella furcifera, Dcit, Diaphorina citri. Amino acid sequences used to construct the tree are shown in Supplementary Table S2. (B) Transcriptional profiles of putative T. papillosa OBPs in different body parts determined by semi-quantitative RT-PCR. The OBPs that are upregulated in the antennae are labeled with red dots. Two reference genes labeled with blue triangle, ubiquitin conjugation factor E4 A (TpapUBE4A) and 60 S ribosomal protein L32 (TpapRPL32), were used as internal references to test the integrity of each cDNA templates. Abbreviations: A: antenna; P, proboscis; SG, stink gland; MG, midgut; T, tarsus. (C) Relative expression levels of antenna-predominant OBPs in the female and male antennae, and other body parts. Abbreviations: FA, female antennae; MA, female antennae; Bo, other body parts. Expression levels were estimated using the 2−ΔΔCT method. The relative expression level is indicated as mean ± SE (n = 4). Standard error is represented by the error bar, and different letters indicate statistically significant difference between tissues (p < 0.05, ANOVA, HSD).
