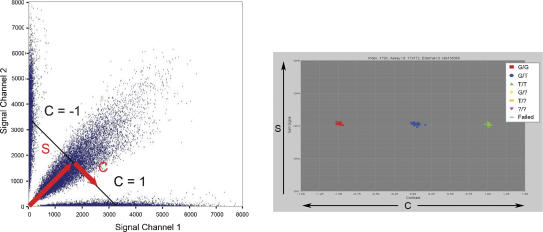
Figure 1.
Calling genotypes based on cluster analysis of raw data. Each SNP in a multiplex assay results in four fluorescent signal values: two for the two expected allele channels and two in background channels. Plotting the signal channels against each other (left) results in the formation of three clusters. The plot on the left shows 50,000 data points across several thousand markers. In order to decouple the overall signal of the particular data point from the contrast between the different allele signals, it is helpful to transform the data into a different space in which the sum of the signals in both channels (S) is plotted on the y-axis and the projection of the individual data point onto the line of constant S (the contrast value C) is plotted on the x-axis. The values of C range from -1 to 1 such that a value of -1 or 1 means signal in only one of the two channels while a value of 0 means equal signal in each channel. A one-dimensional E-M algorithm can then be used to find the clusters of homozygous and heterozygous calls. The colors have been automatically added by the cluster calling algorithm, which has identified the three clusters.