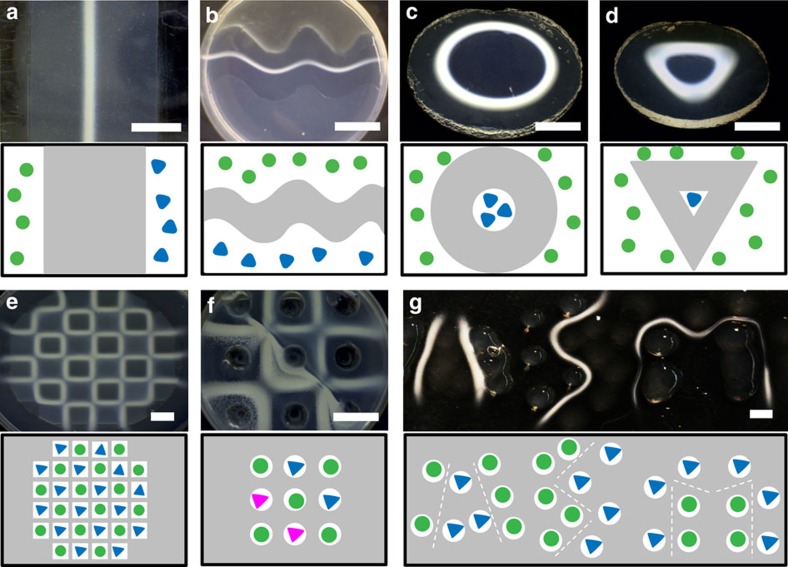
Figure 2. The initial location of reactants controls structure formation in RD-SA.
In a–g, the gel structure formed through RD-SA (photograph, top) resulted from a starting point configuration as shown in the cartoon below. In the cartoons, the grey areas denote the agar matrix, white areas are the reservoirs, which are filled with hydrazide H (green circles) and aldehyde A (blue triangles). (a) A 1D pattern formed by RD of H, from the left, and A, from the right, in an agar diffusion matrix. Method: reservoirs in the agar matrix. (b) By shaping the agar matrix, the RD-SA formed patterns were obtained in different shapes, such as waves. Method: reservoirs in the agar matrix. (c,d) To show the versatility of the RD-formed gel structures, different designs were used to obtain a circle and a triangle. Method: a PDMS mould below the agar matrix. (e) A grid formed by RD-SA. Method: a PDMS mould below the agar matrix. (f) An RD-SA grid made from aldehydes A and A* to compare the difference in SA behaviour, (magenta: A*, blue: A). Method: reservoirs in the agar matrix. (g) Using the RD-SA approach to ‘write’ our research group name (ASM). Method: droplets of reactants in agar solutions were placed on top of an agar matrix, to diffuse and form supramolecular structures at the intersection of the diffusion gradients (Supplementary Methods). Scale bars: 1 cm.