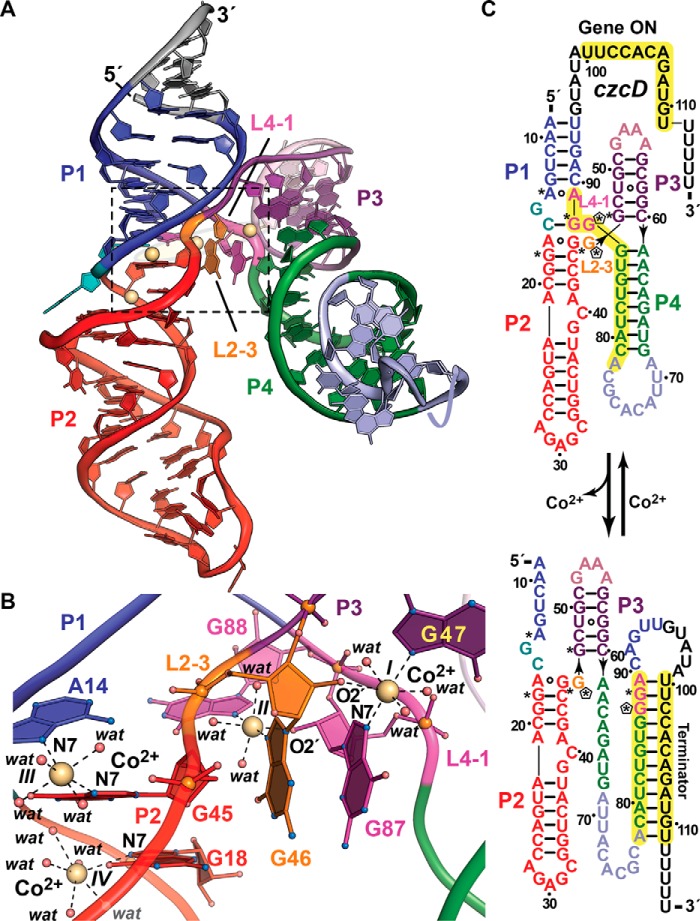
Figure 3.
Tertiary fold and ion coordination by the NiCo riboswitch bound to Co2+. a, ribbon diagram of the Erysipelotrichaceae bacterium riboswitch co-crystal structure determined at 2.64 Å resolution (Protein Data Bank entry 4RUM). The overall architecture comprises tandem, coaxially stacked P1-P2 and P3-P4 helices offset by 30° and joined by a central four-way helical junction. Like the Mg2+-I and Mn2+ riboswitches, divalent ion sensing occurs at the junction interface. Here, four Co2+ sites (I–IV, yellow spheres) bind between loops L2–3 and L4–1. b, close-up view of the divalent ion-binding positions (boxed in a). Sites I–III interact directly with bases G47 and G87, G46 and G88, and A14 and G45, which are conserved among most czc homologs. G46 and G87 each contribute to coordination at sites I and II via purine N7 and ribose hydroxyl groups that form the basis of cooperativity during Co2+ or Ni2+ binding. In contrast to the Mg2+-I and Mn2+ riboswitches that make use of non-bridging phosphate oxygens to coordinate Mg2+ or Mn2+, NiCo riboswitch coordination utilizes one or two N7 moieties, 2′-OH groups, and water molecules to bind Co2+ and presumably Ni2+. c, secondary-structure diagram depicting effector-dependent conformational changes that regulate transcription; asterisks indicate sites of purine N7 Co2+ coordination in b, and pentagons show coordination from ribose 2′-OH groups. Transcriptional regulation by the NiCo riboswitch parallels that of the Mn2+ riboswitch because Co2+ or Ni2+ sensing favors an RNA fold that allows RNA polymerase to read through the transcript, leading to a full-length message that confers heavy-atom resistance when translated. In the effector-free state, P4 remodels to form a terminator hairpin leading to premature transcription termination, potentially resulting in Co2+ or Ni2+ accumulation in the cell.