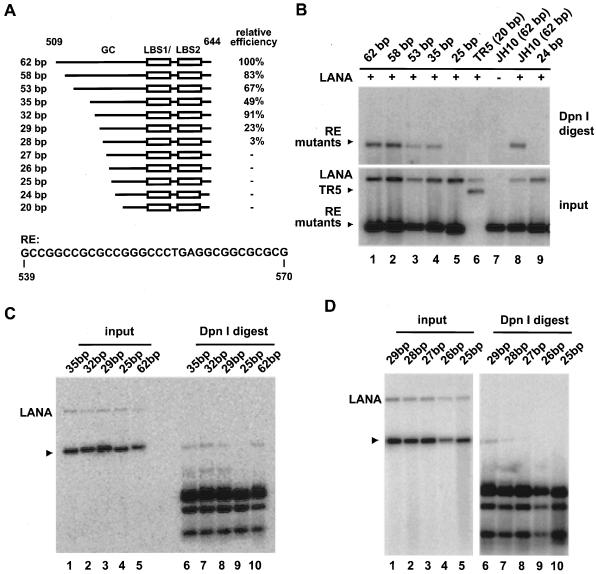
FIG. 2.
Fine mapping of the minimal sequence requirement for RE. (A) Panel of RE mutants. Plasmids contain different lengths of the GC-rich element as indicated upstream of LBS1/2 and 34 bp downstream. Mutants were generated by exonuclease III nuclease digestion of pJH10 (TR nt 509 to 644) or by ligating synthetic oligonucleotides into pBluescript II SK(+). The lower panel shows the RE sequence of 32 bp in length (nt 539 to 570). All replication assays were performed as described for Fig. 1. Replication efficiency was calculated by determining the ratio of input and replicated DNA for each construct in comparison to pJH10, which was set to 100%. Values were derived from two or three independent experiments for each construct. (B) The plasmid containing 35 bp, but not that containing 25 bp, of the GC-rich fragment replicates in the presence of LANA. (C and D) The plasmid containing 29 bp of the GC-rich sequence replicates, the plasmid containing 28 bp has reduced activity, and the plasmid containing 27 bp is not active. Test plasmids are indicated by arrowheads in panels B to D.