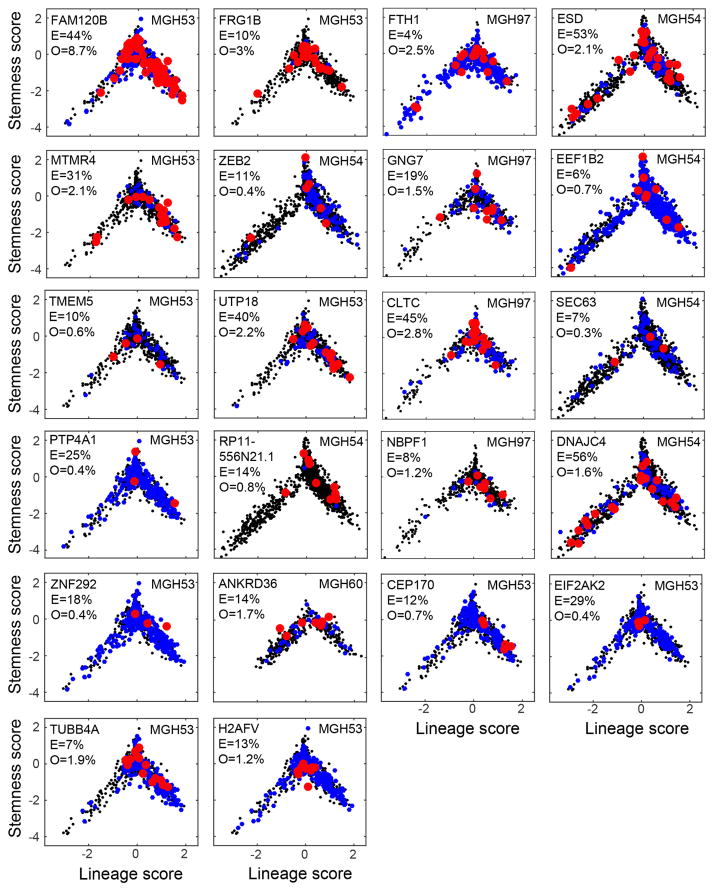
Extended Data Figure 9. Subclonal mutations tend to span the cellular hierarchy.
Each panel shows lineage (x axis) and stemness (y axis) scores of cells in which we ascertained by single cell RNA-seq a mutant (red), a wild-type (blue) or none (black) of the alleles. Included are mutations for which at least three cells were identified as mutants and that were identified by WES as subclonal (fraction < 60%). The gene names, tumour name, ABSOLUTE-derived fraction of mutant cells (E, expected fraction) and the fraction of cells detected as mutant by RNA-seq (O, observed) are also indicated within each panel. We note that identification of a wild-type allele (blue) does not imply a wild-type cell because mutations may be heterozygous, and thus cells could contain both alleles while only one may be detected by single-cell RNA-seq. The observed fraction of mutations (O) is much lower than expected (E) due to limited coverage of the single-cell RNA-seq data, as well as due to heterozygosity. The vast majority of mutations (20 of 22) are distributed across the hierarchy and span multiple compartments. Two remaining mutations (H2AFV and EIF2AK2) appear more restricted to the ‘undifferentiated’ region (intermediate lineage and stemness scores), which could reflect our limited detection rate of mutant cells and/or a bias of the mutation to a particular region. To test the significance of potential biases in the distribution of mutations we calculated, for each mutation, a Euclidean distance among all pairs of mutant cells (based on their lineage and stemness scores), and compared the average pairwise distances among mutant cells to that among randomly selected subsets of the same number of cells. None of the mutations were significant with a false discovery rate (FDR) of 0.1, although this could reflect our limited statistical power and we cannot exclude a potential bias. The apparent bias of mutant cells to the OC lineage over the AC lineage (that is, positive versus negative lineage scores) reflects the lower frequencies of AC-like cells compared to OC-like cells in MGH53 and MGH54 (MGH53: 17% AC versus 39% OC; MGH54: 23% AC vs. 45% OC); this bias is also observed for the detection of wild-type alleles (blue) demonstrating that there is no bias against mutation detection in the AC lineage.