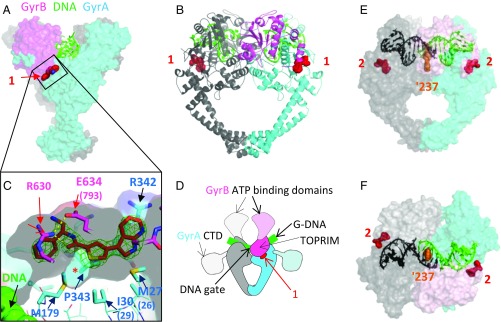
Fig. 3.
Crystal structures of compounds 1 and 2 with S. aureus DNA gyrase and DNA. (A) Compound 1 binds in a pocket between the GyrA (cyan) and GyrB (pink) subunits in the complex with DNA (green) and S. aureus GyrB27A56. (B) Orthogonal view of complex with protein shown in schematic and one GyrB-GyrA subunit in pink and cyan and the other in gray. Carbon atoms of compound 1 are in red. (C) An enlarged view of the ligand binding site looking from GyrB (R630 and E634 are near the viewer). A semitransparent surface on the protein is shown gray on the inside (view from GyrB) and colored on the outside. Note that the front clipping plane cuts through the near surface at one point—allowing a clearer view of the “hydrophobic cavity” under the compound (indicated with *). GyrA P343 is behind the far surface of the pocket. An Fo-Fc ligand omit map at 3.5 sigma (green mesh) is shown. Note that DNA is remote from the compound binding site. (D) Orientation schematic showing the location of the compound 1 binding site (same view and color scheme as in B). The dashed delineated domains are absent from the crystallographic core construct, and their approximate orientation is indicated. (E and F) Two orthogonal views of the 2.22-Å crystal structure of compound 2 with S. aureus DNA gyrase, DNA, and GSK945237 (orange spheres, an NBTI used to cocrystallize and aid crystallography) (Materials and Methods). The protein subunits are shown with semitransparent surface so that the thiophene (red spheres) can be seen. The DNA is colored black/green to indicate the positions of the four base pair staggered breaks.