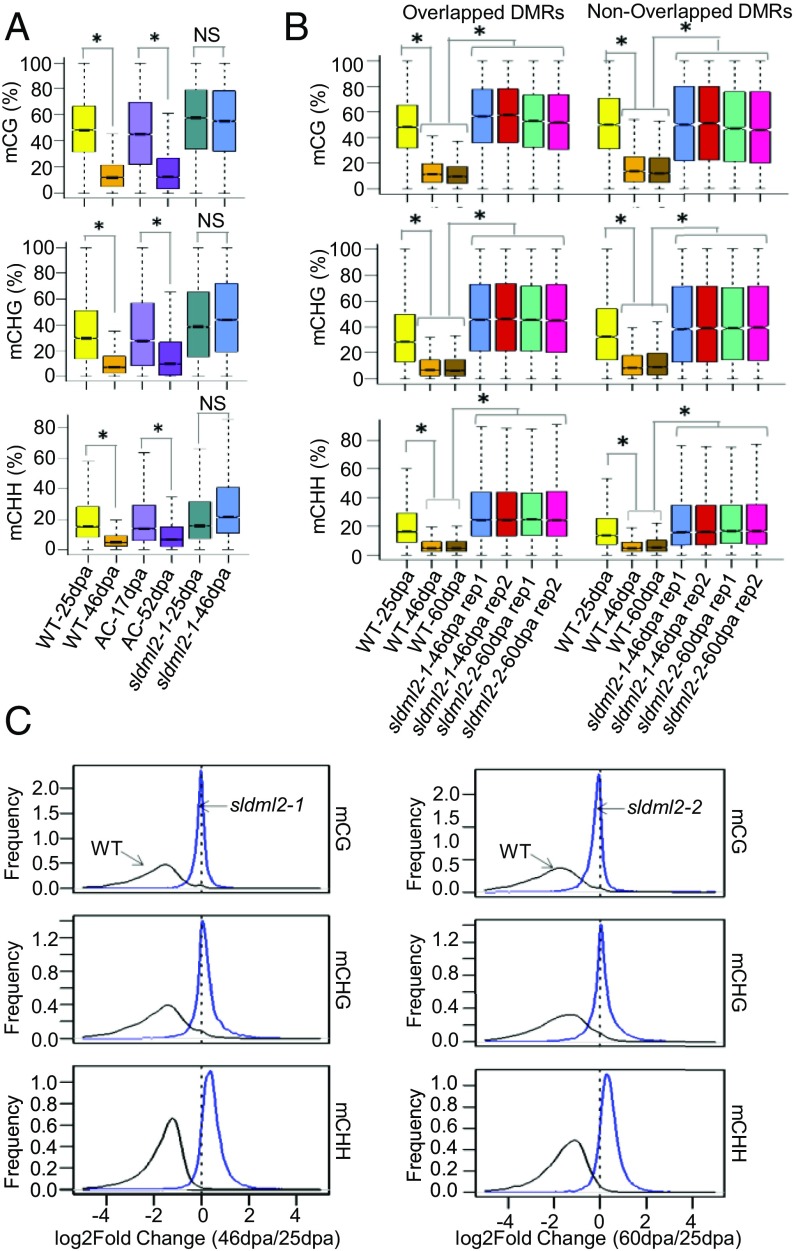
Fig. 4.
SlDML2 is required for ripening-induced DNA demethylation. (A) Boxplots showing mCG, mCHG, and mCHH levels at ripening-induced hypo-DMRs in WT-25dpa, WT-46dpa, AC-17dpa, AC-52dpa, sldml2-1-25dpa, and sldml2-1-46dpa (average of two replicates) (*P value < 2.2e−16, one-tailed t test; NS, not significant). (B) Boxplots showing mCG, mCHG, and mCHH levels at ripening-induced hypo-DMRs that are overlapped (Left) or not overlapped (Right) with sldml2-1 hyper-DMRs in WT-25dpa, WT-46dpa, WT-60dpa, two biological replicates of sldml2-1-46dpa, and two biological replicates of sldml2-2-46dpa. For mCG, mCHG and mCHH levels, WT-25dpa, two biological replicates of sldml2-1-46dpa, and two biological replicates of sldml2-2-46dpa were significantly higher than WT-46dpa and WT-60dpa (*P value < 2.2e−16, one-tailed t test). (C) The distribution of logtwofold-change (46dpa/25dpa) of mCG, mCHG, and mCHH levels in sldml2-1 mutant and the WT (Left), and the distribution of logtwofold-change (60dpa/25dpa) in the sldml2-2 mutant and the WT (Right). A peak at 0 corresponds to no change in DNA methylation during ripening.