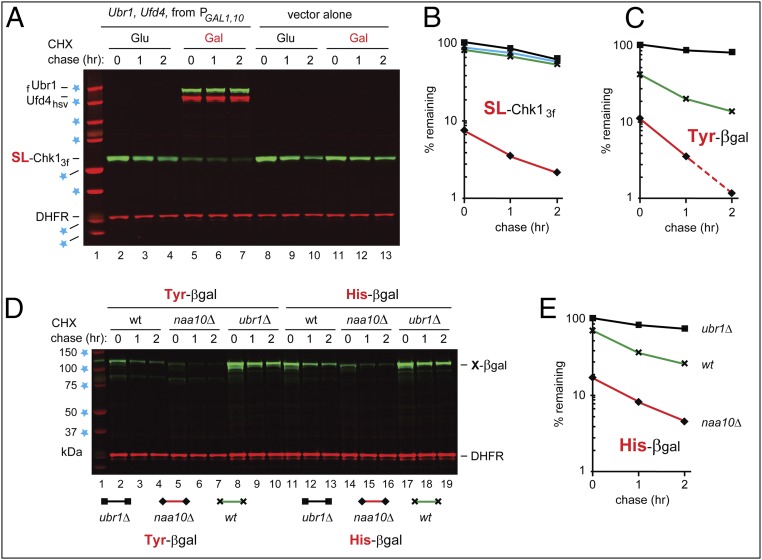
Fig. 5.
Overexpression of the Arg/N-end rule pathway destabilizes Chk1. (A) CHX-chases, using PRT (Fig. 2B), with wild-type SL-Chk13f, for 0, 1, and 2 h, in wild-type S. cerevisiae carrying a high-copy plasmid that expressed both flag-tagged Ubr1 (fUbr1) and Hsv-tagged Ufd4 (Ufd4hsv) from the bidirectional PGAL1,10 promoter. Lanes 2–4 and 5–7: cells containing the PGAL1,10-UBR1/UFD4 plasmid in glucose and galactose medium, respectively. Lanes 8–10 and 11–13 are as in lanes 2–4 and 5–7, respectively, but with cells containing a vector alone. The presence of fUbr1 and Ufd4hsv in lanes 5–7 was verified directly, using anti-flag and anti-Hsv antibodies. Blue stars denote 20-, 25-, 37-, 50-, 75-, 100-, 150-, and 250-kDa markers. (B) Quantification of data in A. Rhombuses, squares, triangles, and crosses denote, respectively, the decay curves of SL-Chk13f in wild-type cells overexpressing Ubr1-Ufd4 (in galactose), or containing vector alone (in glucose), or containing the repressed Ubr1-Ufd4 plasmid (in glucose), or containing vector alone (in galactose). (C) Quantification of data in D, lanes 2–10. For curve designations, see the keys below the immunoblot in D. (D) Lane 1, kDa markers. CHX-chase, using PRT (Fig. 2B), for 0, 1, and 2 h, with wild-type (lanes 2–4), naa10Δ (lanes 5–7), and ubr1Δ (lanes 8–10) S. cerevisiae expressing Tyr–β-gal, derived from Ub–Tyr–β-gal. Lanes 11–19 are as in lanes 2–10 but with His–β-gal, derived from Ub–His–β-gal. Blue stars indicate 37-, 50-, 75-, 100-, and 150-kDa markers. (E) Quantification of data in D, lanes 11–19. For curve designations, see the keys below the immunoblot in D. All chases in this study were performed at least twice, yielding results that differed by less than 10%.