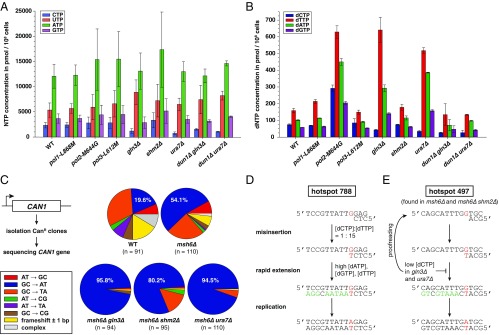
Fig. 4.
Inactivation of Gln3 or Ura7 results in NTP and dNTP imbalance causing increased G:C-to-A:T transitions. (A) NTP and (B) dNTP pool measurements in the indicated strains (Tables S4 and S5). Error bars represent SD. (C) Independent CanR clones (n ≥ 91 per genotype) were sequenced for CAN1 mutations. Graphs indicate the type of identified mutations, in percentages (see also Tables S6 and S7). (D) The G-to-A mutational hotspot at nucleotide 788 was frequently found in msh6Δ gln3Δ, msh6Δ shm2Δ, and msh6Δ ura7Δ strains. Predicted mutation is noted in red. Nucleotides marked in green are facilitating mispair rapid extension prior proofreading due to their higher abundance in gln3Δ or ura7Δ mutants, compared with the WT strain. (E) The G-to-A mutational hotspot at position 497 was frequently found in msh6Δ and msh6Δ shm2Δ, but not in msh6Δ gln3Δ and msh6Δ ura7Δ. Here, the predicted G-dT mispair is immediately followed by incorporation of dCTP (in black), which is less abundant in gln3Δ and ura7Δ strains and, therefore, unlikely to support mispair rapid extension.