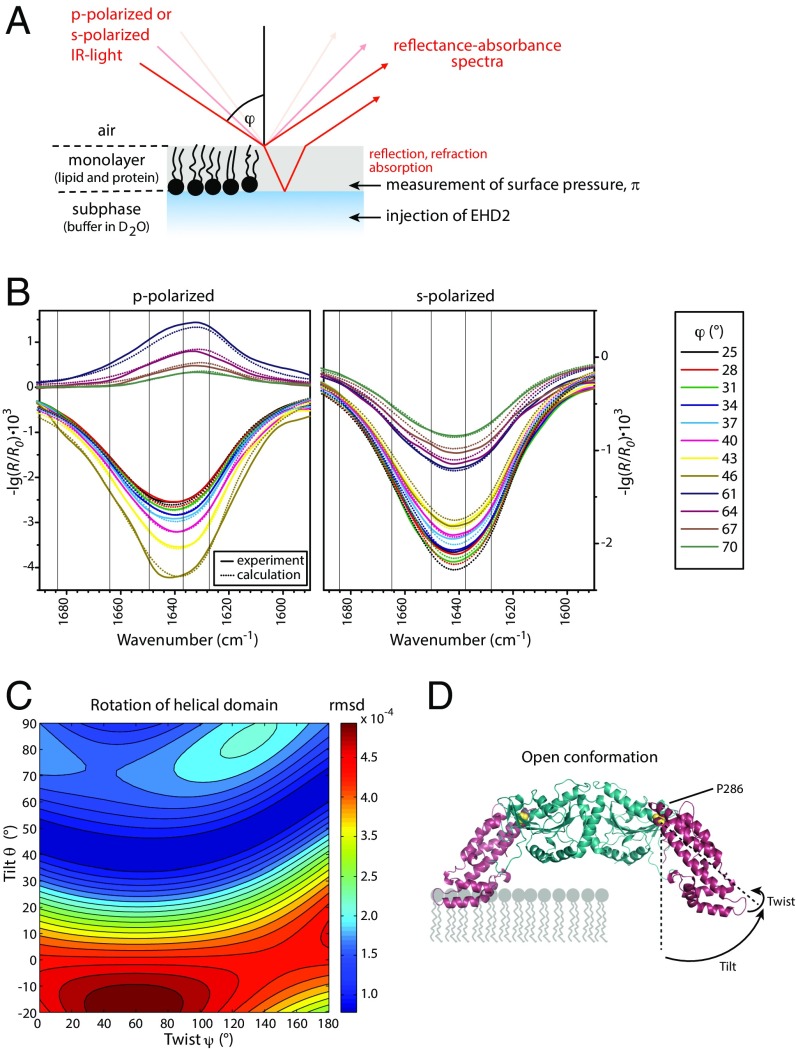
Fig. 3.
EHD2 binds to lipid membranes in an open conformation as determined by IRRAS. (A) Scheme of the IRRAS setup: polarized IR light is reflected at the air–water interface covered by the lipid monolayer including adsorbed proteins. The angle of incidence φ is varied from 25° to 70° in steps of 3°. The reflected IR beam is detected and converted to reflectance–absorbance spectra of the surface film. (B) Sets of IRRA spectra of EHD2 ΔEH preincubated with AMP-PNP and adsorbed to a lipid monolayer, shown in the spectral region of the amide I′ vibrations. The spectra are recorded with IR light in p-polarization (Left) and s-polarization (Right) and at different incidence angles φ (see legend). Experimental spectra (solid lines) are shown together with calculated spectra (dotted lines) of EHD2 ΔEH in open conformation, that is, with the helical domains rotated outward (θ = 44°, ψ = 40°). (C) Color-coded error map of cumulative root-mean-square deviation (rmsd) of calculated versus experimental IRRA spectra as function of the tilt angle θ and twist angle ψ of the helical domain. A good accordance between experimental and calculated spectra is colored blue and thereby denotes probable orientations of the helical domains in membrane-bound EHD2 ΔEH. The spectra shown in B result in a minimal rmsd in this error map. A bad accordance between experimental and calculated spectra is colored red and depicts improbable conformations of EHD2 ΔEH bound to lipid monolayers. Note that the crystallized conformation (PDB ID code 4CID) (θ = 0; ψ = 0) is an improbable conformation of the membrane-bound state of EHD2. (D) Predicted conformation of EHD2 ΔEH bound to the lipid monolayer with illustration of tilt angle θ and twist angle ψ of the helical domains (purple) as used in the spectra calculations. The tilt angle is the angle between the helical domain main axis (dotted line) with respect to the monolayer normal. The twist angle is the angle of rotation around the helical domain main axis. Helical domain and G domain are linked at proline 286 (yellow), which is defined as the center of rotation for helical domain tilting. Monolayer lipids are depicted schematically (gray) in scale to the protein.