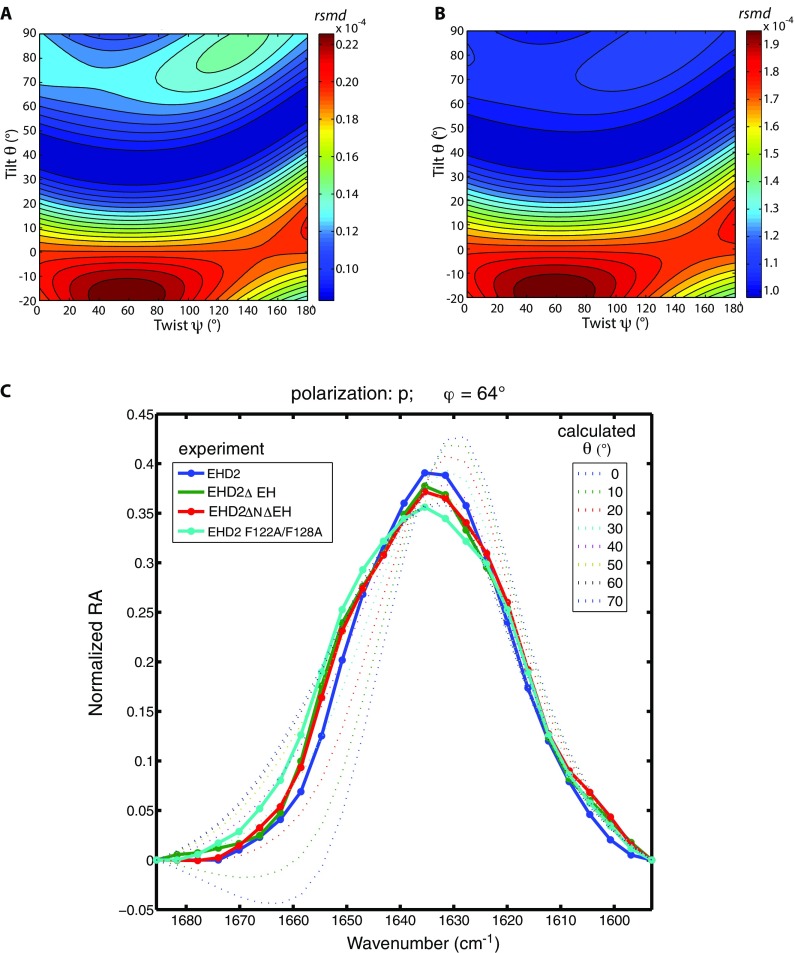
Fig. S6.
The conformational analysis is robust, and all investigated mutants of EHD2 as well as the wild type bind lipid monolayers in an open conformation. (A and B) Root-mean-square deviation (rmsd) of calculated versus experimental IRRA spectra after vector normalization of each spectrum (A) and rmsd of the experimental and calculated dichroic ratio spectra (RAp/RAs) (B). The rmsd is shown as color-coded contour map in dependence of the helical-domain tilt angle θ and twist angle ψ. In these maps, the same data are compared as in Fig. 3C (main text), but their similarity to the calculated spectra is defined in different ways. By normalizing the spectra (A) before rmsd calculation, the comparison is based on the band shapes and the intensity information is dismissed. By calculating the rmsd of the dichroic ratios (B), the spectra recorded in s-polarization contribute more to the rmsd than in the analysis of cumulative rmsd shown in Fig. 3C, and the band shape of the spectra measured in p-polarization becomes less important. Note that all different ways of comparing calculated spectra to experimental spectra lead to the same conclusion that the helical domains of EHD2 ΔEH rotate outward (θ = 40–70°) upon binding of the protein to the lipid monolayer. This proves that the developed method of conformational analysis is robust to the way the rmsd is defined. (C) The detailed conformational analysis based on IRRAS was performed on EHD2 ΔEH, but also the other mutants discussed in this report were investigated by IRRAS. To assess their conformation when bound to a lipid monolayer after preincubation with AMP-PNP, we present their vector-normalized spectra measured with p-polarized IR light at an incidence angle of φ = 64° in the amide I′ spectral region (solid lines with symbols). At this specific polarization and incidence angle, the band shape of the amide I′ band is most sensitive to reorientation of the helical domain. All experimental spectra show a similar amide I′ band shape, suggesting a similar protein conformation. Along with the experimental spectra, normalized spectra calculated for various orientations of the EHD2 helical domain (dotted lines) are shown. The twist angle is ψ = 40° in all calculations. The helical-domain tilt angle is systematically increased from θ = 0° to θ = 70° for calculating the spectra. Note that all experimental spectra compare well to spectra calculated with a helical-domain tilt of θ = 30° to 60°, that is, an open conformation of EHD2.