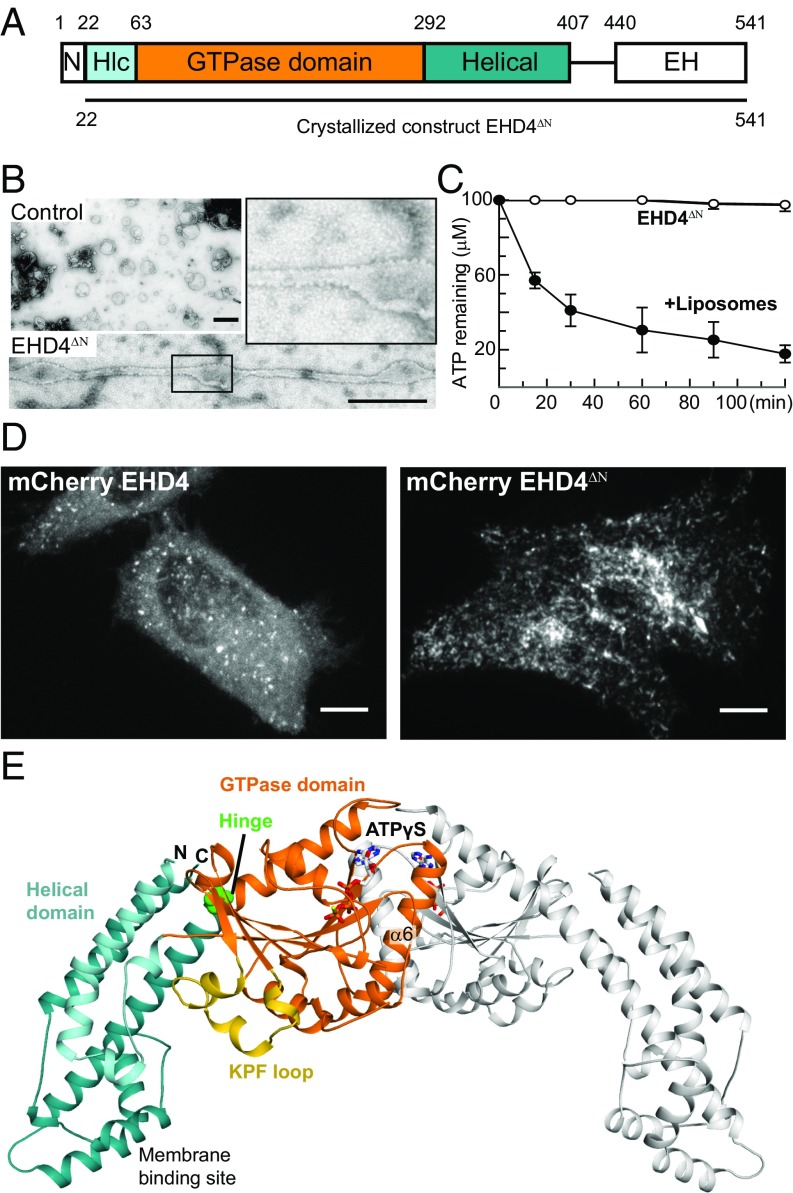
Fig. 1.
Structure of the activated EHD4 dimer. (A) Domain architecture of EHD proteins. Numbers refer to the amino acid sequence of mouse EHD4. (B) EM micrographs of negatively stained EHD4ΔN in the presence of liposomes. As a control, liposomes were stained without the addition of EHD4ΔN. (Scale bars: B, 500 nm; D, 10 μm.) (C) ATPase assays of EHD4ΔN in the absence and presence of Folch liposomes were carried out at 30 °C. Error bars represent the range of two independent measurements. (D) EHD4 and EHD4ΔN were expressed in HeLa cells with a C-terminal mCherry tag. (E) Structure of the ATPγS-bound EHD4ΔN. In the left molecule, domains are colored according to the domain architecture, the conserved hinge around Pro289 is indicated in green. The right molecule is shown in gray. Note that the EH domains were not resolved in the electron density.