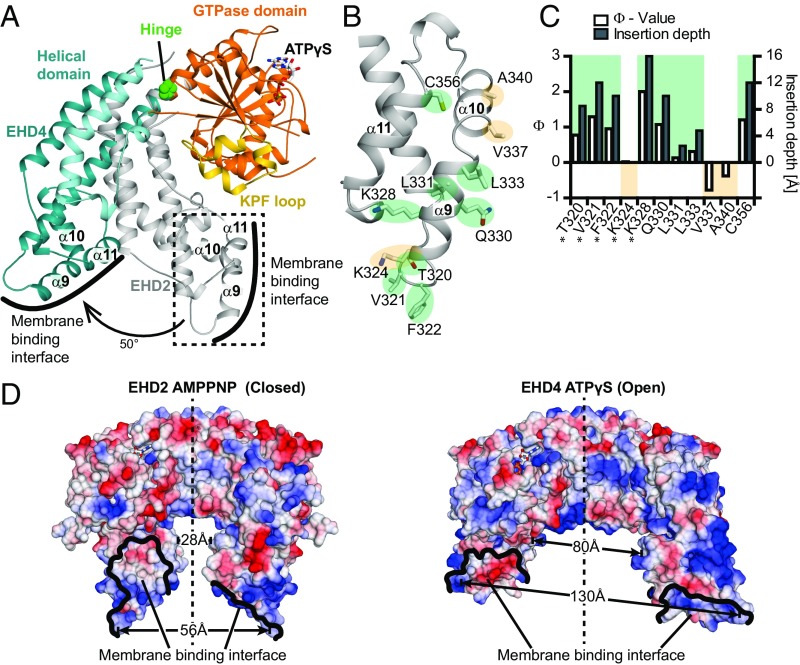
Fig. 2.
A rotation of the helical domains allows membrane binding. (A) Superposition of the GTPase domains of ATPγS-bound EHD4ΔN and AMPNP-bound EHD2 (PDB ID code 4CID, only the helical of EHD2 is shown). Pro289 acts as the hinge for a 50° rotation of the helical domain. (B) Magnification of the boxed area in A showing details of the membrane binding interface of EHD2. Membrane inserting (green)/noninserting (yellow) residues are highlighted. (C) The logarithmic ratio Φ of the accessibilities of spin labels to the paramagnetic colliders O2 and NiEDDA was calculated for EHD2 labeled at the indicated positions in the presence of Folch-SUVs (open bars referring to the left y axis). Results from residues 277, 320–324, and 328 (*) are from ref. 16. Positive Φ values indicate membrane insertion based on prior calibration with spin-labeled lipids. This calibration was used to convert Φ values into membrane insertion depth of each residue (filled bars to the right y axis). (D) Electrostatic potentials (±10 kcal/mol × e, where e is the charge of an electron) were plotted on the surfaces of the EHD2 (Left) and EHD4ΔN (Right) dimers, with the GTPase domains of both dimers in the same orientation. The distances between Phe322-Phe322 in EHD2 (56 Å), Phe325-Phe325 in EHD4 (130 Å), Pro350-Pro350 in EHD2 (28 Å), and Ala353-Ala353 in EHD4 (80 Å) are indicated. The membrane binding surface of EHD2 is more positively charged compared with EHD4, possibly reflecting different lipid binding specificities (Fig. S4).