Fig. S1.
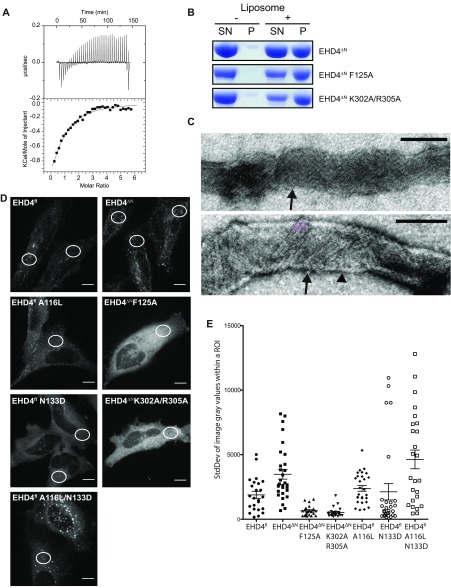
Functional characterization of EHD4ΔN. (A) In ITC experiments, a 2 mM ATPγS solution was titrated into a 70 µM EHD4ΔN solution. The resulting heat changes were integrated and fitted to a quadratic binding model. A KD of 77 µM, with a binding number n of 0.67 was obtained. Note that this experiment was carried out at 500 mM NaCl, which was required to prevent protein precipitation. The high salt concentration may negatively influence the binding affinity. (B) Folch liposome cosedimentation assays of EHD4ΔN and the indicated mutants. P, Pellet fraction; SN, supernatant. (C) Liposome tubulation experiments as in Fig. 1B. Some tubulated liposomes (arrowhead) contained a regular EHD4 coat with striations of 9 nm width (black arrows), which corresponds to the width of our EHD4 oligomeric model (Fig. 4A). (D) Representative maximum intensity projections of live HeLa cells expressing mCherry-tagged protein as indicated. White circles illustrate typical placings of the region of interest (ROI) within the cells. (E) SD of gray values derived from one ROI per cell from at least 20 cells for each indicated construct. Error bars show SEM. Two-tailed t tests was performed on EHD4ΔN to each set of data to determine significance *P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0001. (Scale bars: 10 μm.)