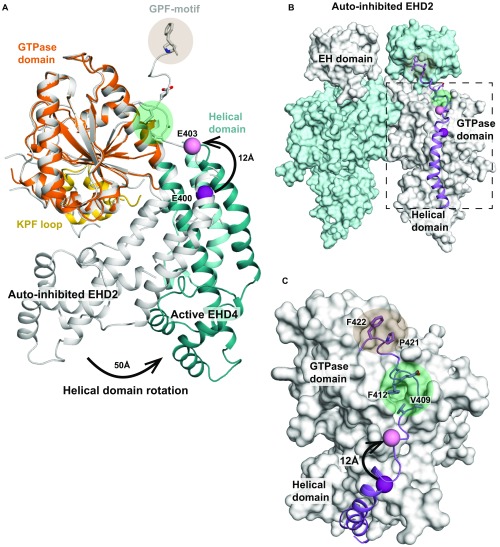
Fig. S6.
Reorientation of the linker in the active conformation. (A) The GTPase domains of EHD2 and EHD4ΔN were superimposed. Highlighted is the position of Glu403 (represented as pink ball), the last resolved residue in the EHD4ΔN structure, corresponding to Glu400 in EHD2 (represented as purple ball). This residue is displaced by 12 Å through the rotation of the helical domain. (B) Autoinhibited EHD2 dimer (PDB ID code 4CID), in which the final helix α12 and the adjacent linker is shown in purple. (C) Detailed view showing the interaction of the helical domain-EH domain linker with the GTPase domain. The displacement of Glu400/Glu403 is indicated as in A. We suggest that the rotation pushes the linker away from its position in the autoinhibited state EHD2 dimer. In particular, Val409 and Phe412 will be shifted away from their hydrophobic binding pocket (marked as green circle). Consequently, also the GPF motif (residues 420–422, marked as orange circle), which binds to the opposing EH domain, may be displaced.