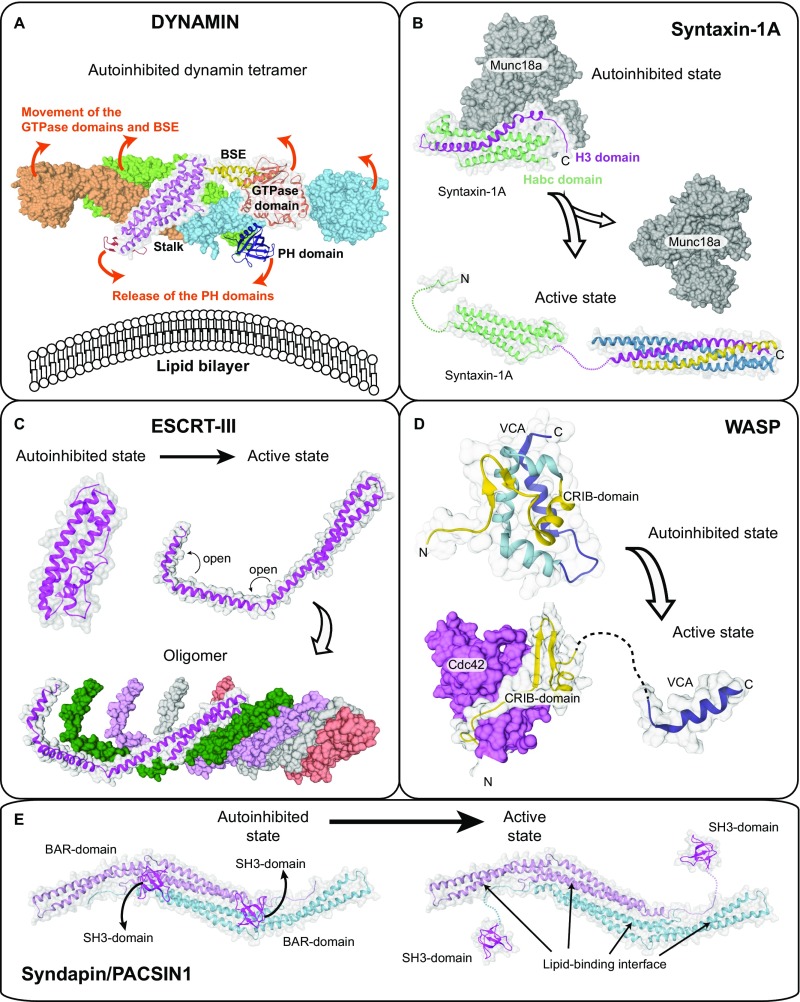
Fig. S8.
Activation mechanism of other membrane-associated proteins. (A) Crystal structure of the autoinhibited dynamin-3 tetramer (PDB ID code 5A3F). Based on a low-resolution EM reconstruction of oligomerized dynamin, it was suggested that the PH domains are released from the autoinhibitory stalk interface during activation and bind to the membrane, whereas the GTPase domains dimerize via the G-interface across helical turns (suggested movements are indicated by arrows). The PH domain release is thought to expose oligomerization interfaces in the stalk, therefore permitting oligomerization. Because there is no high-resolution structure of dynamin in the oligomerized state available, the exact mechanistic details of activation are still elusive. (B) The SNARE subunit Syntaxin-1A forms a closed autoinhibited four helix-bundle conformation (PDB ID code 3C98) and an open three helix-bundle conformation (comprising the “Habc domain,” PDB ID code 1S94). In the open state, the released H3 helix (magenta) associates with other SNARE partners to form the assembled SNARE complex. Binding of the regulatory Munc18a/Sec1-subunit stabilizes the autoinhibited conformation of syntaxin1A. (C) The ESCRTIII-component CHMP1B in the closed (modeled based on PDB ID code 3GGY) and open state (based on an electron microscopy reconstruction of the IST1-CHMP1B ESCRT-III copolymer, PDB ID code 3JC1). Domain opening exposes oligomerization interfaces in CHMP1B and allows assembly of a heteromeric scaffold. (D) Autoinhibited (PDB ID code 1EJ5) and active conformation (PDB ID code 1CEE) of the WASP protein. In the autoinhibited state, the CRIB domain tightly interacts with the VCA peptide. The small GTPase Cdc42 binds to the CRIB domain, therefore releasing the VCA peptide to induce actin polymerization. Domain opening is further promoted by PIP2 binding. (E) Crystal structure of the autoinhibited Syndapin/PACSIN-1 dimer comprising BAR and SH3 domains (PDB ID code 2X3W). Only one SH3 domain was observed in the crystal structure; the localization of the second SH3 domains was modeled. Upon binding to proline-rich sequences of target proteins such as dynamin, the SH3 domains are released, therefore exposing the membrane binding site in the BAR domain to allow formation of a membrane-associated BAR domain scaffold. Similar to EHDs, these peripheral membrane proteins show a closed, autoinhibited conformation and an active open conformation. In the open conformation, self-assembly (dynamin, ESCRT-III), membrane binding (dynamin, PACSIN-1, ESCRT-III, WASP) or protein–protein interaction motifs (all examples) become exposed. However, the complex coupling of membrane binding to domain releases, rotations, and loop switches is specific for EHDs and possibly other dynamin proteins.