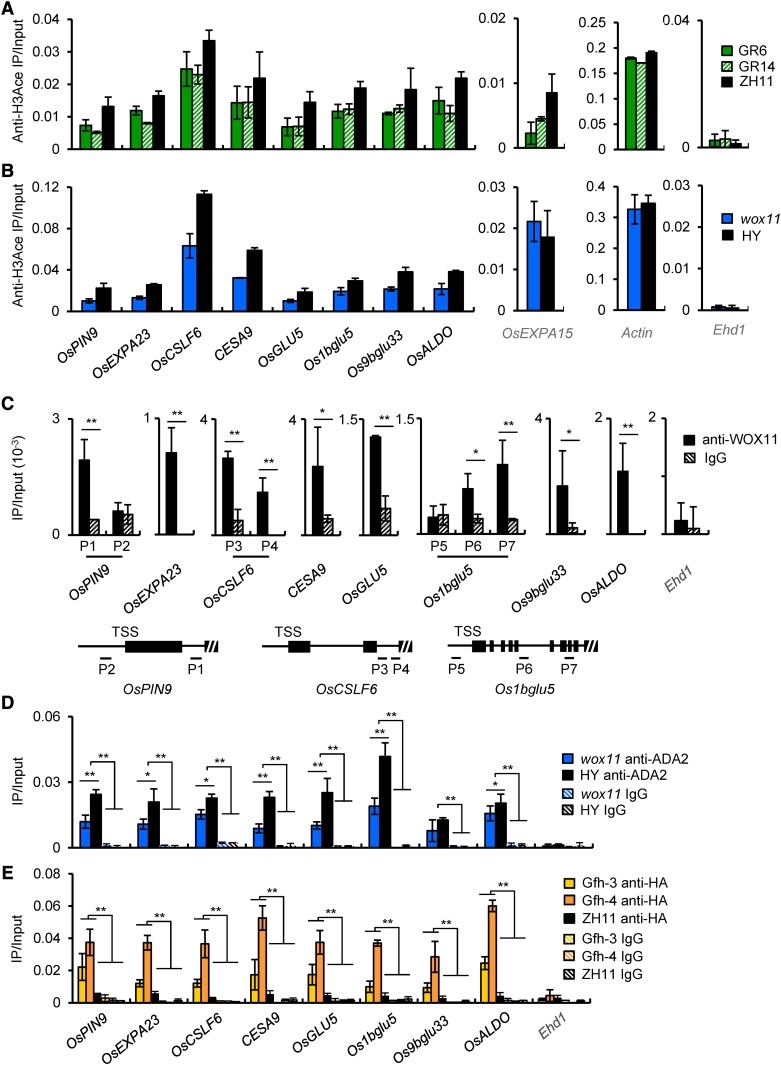
Figure 7.
ADA2-GCN5 and WOX11 Directly Bind to Common Downstream Genes and Regulate Histone H3 Acetylation in Crown Root.
(A) and (B) GCN5 and WOX11 regulate histone H3 acetylation of eight common downstream genes. Histograms on left are ChIP assays with anti H3ace of eight common target genes in roots of two GCN5 RNAi lines compared with the wild type (ZH11) (A) and wox11 mutant compared with the wild type (HY) (B). Histograms on side are tests of OsEXPA15 and ACTIN loci as controls. Bars represent means ± sd from three technical repeats of one biological replicate. The other two biological replicates are shown in Supplemental Figure 9. Each biological replicate was performed using crown roots collected from independently germinated seedlings of each genotype.
(C) WOX11 binds directly to common target genes. ChIP assays were performed using anti-WOX11 antibody with roots of wild-type plants. Anti-IgG was used as a control. Three of the eight genes have more than one putative WOX binding site. Primer sets corresponding to each of the binding sites were used (see schematic below the graphs).
(D) and (E) ADA2 and GCN5 associate directly with the downstream genes. ChIP assays were performed using polyclonal anti-ADA2 (D) or anti-HA (E). Anti-IgG was used as a control. ChIP assays with anti-ADA2 or anti-HA antibodies were performed with root chromatin fragments of wox11 mutant and wild-type (HY) plants and of GCN5-falg-HA and wild-type (ZH11) plants, respectively.
Bars in (C) to (E) are means ± sd from three biological replicates. Each replicate was performed using crown roots harvested from independently germinated seedlings for each genotype. Significant enrichments (Student’s t tests) calculated from three biological replicates are indicated: *P value < 0.05 and **P value < 0.01.