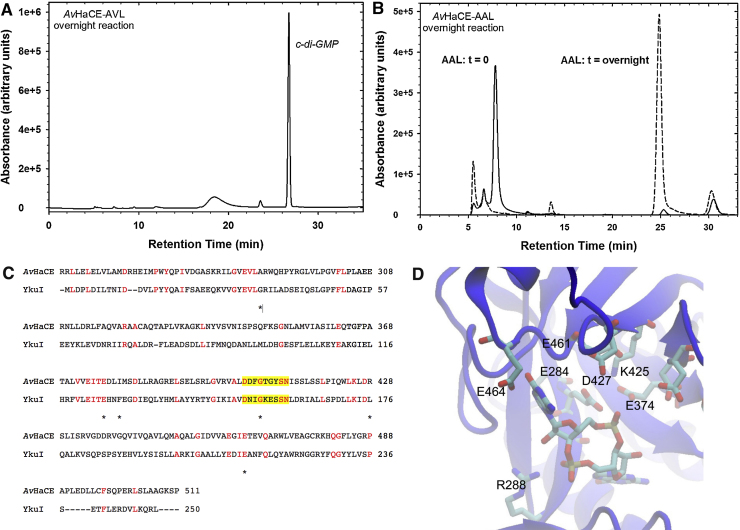
Fig. 2.
Residues important for phosphodiesterase activity. (A) HPLC analysis of the reaction products of the AVL variant of AvHaCE after incubation overnight. (B) HPLC analysis of the reaction products of the AAL variant of AvHaCE before and after incubation overnight. The HPLC analysis of the nucleotide mixture immediately after adding AvHaCE is also illustrated, indicting the retention time of GTP. (C) Primary structure alignment of AvHaCE and the phosphodiesterase YkuI from B. subtilis (BSU14090). Conserved residues are highlighted in red. Residues shown to be important for phosphodiesterase activity of RocR from P. aeruginosa[42] are indicated by the asterisk. The yellow highlight indicates the position of loop D. (D) Homology model of residues 249-502, corresponding to the phosphodiesterase domain, of AvHaCE. The model was built with YkuI (pdb 2W27) as the template. Select amino acids proposed to be located within the phosphodiesterase active site of AvHaCE, as well as bound cyclic-diGMP, are rendered in stick format. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)