Abstract
A control point for keeping redox homeostasis in Saccharomyces cerevisiae during fermentative growth is the dynamic regulation of transcription for the glycerol-3-phosphate dehydrogenase 2 (GPD2) gene. In this study, the possibility to steer the activity of the GPD2 promoter was investigated by placing it in strains with different ability to reoxidise NADH, and applying different environmental conditions. Flow cytometric analysis of reporter strains expressing green fluorescent protein (GFP) under the control of the GPD2 promoter was used to determine the promoter activity at the single-cell level. When placed in a gpd1Δgpd2Δ strain background, the GPD2 promoter displayed a 2-fold higher activity as compared to the strong constitutive glyceraldehyde-3-phosphate dehydrogenase (TDH3). In contrast, the GPD2 promoter was found to be inactive when cells were cultivated in continuous mode at a growth rate of 0.3 h−1 and in conditions with excess oxygen (i.e. with an aeration of 2.5 vvm, and a stirring of 800 rpm). In addition, a clear window of operation where the gpd1Δgpd2Δ strain can be grown with the same efficiency as wild type yeast was identified. In conclusion, the flow cytometry mapping revealed conditions where the GPD2 promoter was either completely inactive or hyperactive, which has implications for its implementation in future biotechnological applications such as for process control of heterologous gene expression.
Keywords: GPD2 promoter, Inducible promoter, Glycerol-deficient yeast, GPD deletion, Flow cytometry, Redox reporter, NADH/NAD+, Population heterogeneity, Recombinant protein production
1. Introduction
Baker’s yeast, Saccharomyces cerevisiae, can be used for production of a range of different commodity chemicals [1], [44], biofuels [20], [25], [41], bioflavours [8], fine chemicals [45], [46], [54] and pharmaceuticals [17], [53]. It is used as production host for approximately 20% of biopharmaceuticals approved by the Food and Drug Administration (FDA) or by the European Medicines Agency (EMA) [15]. In all yeast-based processes, a carbon source, which is normally sugar, is converted into biomass, and depending on the specific process, also into a spectrum of products and by-products. To reach optimal productivity and yield of biomass and products, it is often crucial to control the central metabolic pathways for efficient carbon assimilation and reduction of by-products. Not least control of NADH/NAD+-ratios during processing can be important since it determines the thermodynamic feasibility of a large number of metabolic reactions [16], whereof some are more beneficial than others from a process perspective.
Glycerol is a preferred metabolic end-product during winemaking where it contributes favourably to the organoleptic characteristics of the product [10]. However, in most other cases, glycerol is a yield-lowering by-product that negatively influences process economy [55]. From an applied perspective, there is thus a need to monitor and control the requirement for glycerol formation, or perhaps more importantly the need for NADH oxidation in the production strain and during different process conditions. Production of glycerol by yeast under fermentative conditions occurs as a consequence of a need for recycling of NADH formed from anabolic reactions, such as amino acid biosynthesis [32], [51]. The gene products of glycerol-3-phosphate dehydrogenase (GPD) catalyse the NADH-dependent conversion of dihydroxyacetone phosphate (DHAP) to glycerol-3 phosphate, which is further converted to glycerol by dl-glycerol-3-phosphatase [2], [11], [33]. The GPD activity resulting from the two isoenzymes Gpd1p and Gpd2p is essential for growth under anaerobic conditions; however, removal of one of the isoenzymes by gene deletion can be compensated by the activity of the other [3]. Under aerobic conditions the double gene knockout has previously been reported to be able to grow, but at a reduced rate compared to wild-type yeast [6].
The GPD2 promoter activity is known to be up-regulated during growth under conditions with limited supply of oxygen, i.e. under conditions with elevated need for fermentative oxidation of NADH [3]. However, a thorough investigation of its relative activity in strains with different capacity for anaerobic NADH oxidation, and during growth at different dilution rates in continuous cultivation mode and under different levels of aeration is still lacking. Furthermore, the potential of GPD2p for controlling recombinant gene expression by varying environmental conditions has previously not been addressed. The controlling of promoter activity is a useful tool to optimize activities of heterologous biosynthetic pathways. Recombinant gene expression in S. cerevisiae is often made from constitutive native promoters of different strengths [36], [47]. There are also several inducible promotors, such as CUP1p (copper-inducible promoter)[12], that previously have been employed to steer recombinant gene expression. In recent years, screening of synthetic promoter libraries has been used to identify promoters with varying activity under different applied conditions (see for example review by Redden et al. [39].
Flow cytometry (FC) analysis of green fluorescent protein (GFP) based biosensors is a well-established and rapid methodology for determining promoter activity in microbial populations (reviewed by Fernandes et al. [14]. The usefulness of FC in combination with fluorescent proteins has previously been demonstrated for measuring the activity of a number of different yeast expression systems [24], [47]. Also, FC have been used to measure fluorescent proteins correlated to other user-specified properties, for example for measuring intracellular pH [49] or for monitoring cell growth and cell membrane robustness [7].
In this study, a set of previously developed reporter strains [26] with or without GPD activity, where expression of green fluorescent protein (GFP) is under control of the GPD2 promoter, were cultivated under different conditions in batch, accelerostat, and chemostat mode and at different levels of aeration. Flow cytometric analysis was employed to determine a blueprint of the GPD2 promoter activity within the cell population with single-cell resolution. Our results suggest that if sufficient oxygen is supplied the GPD2 promoter is fully inactive in both glycerol-positive (wild-type background) and glycerol-negative (gpd1Δgpd2Δ background) yeast as long as the growth rate is controlled at approximately 0.3 h−1. However, for a gpd1Δgpd2Δ strain grown at maximum growth rate in batch cultivation mode with limited oxygen supply, the activity of the GPD2 promoter was higher than the well-known strong constitutive glyceraldehyde-3-phosphate dehydrogenase (TDH3) promoter. The potential of using the GPD2 promoter for recombinant gene expression is discussed, including the possibility to induce its activity by simply controlling process parameters such as level of aeration and substrate feed-rate.
2. Material and methods
2.1. Strains
The S. cerevisiae strains used in the study are summarized in Table 1. Escherichia coli NEB 5α (New England BioLabs, Ipswich, MA, USA) was used for subcloning. All strains were stored in 20% glycerol at −80 °C. Yeast cells from freshly streaked yeast nitrogen base (YNB) plates (6.7 g/l yeast nitrogen base without amino acids, 20 g/l glucose, and 2% agar) were used for all cultivations.
Table 1.
Strains and plasmids used in this study
| Strains and plasmids | Relevant features | Reference |
|---|---|---|
| Plasmids | ||
| p426GPD | URA3 Mumberg vector | [29] |
| pYGFP3 | URA3 ADH1p-yEGFP3-ADH1t | [9] |
| YIpJK01 | HIS3 GPD2p-yEGFP3-PGK1t | [26] |
| YEpJK01 | URA3 GPD2p-yEGFP3-PGK1t | [26] |
| YEpJK03 | URA3 TDH3- yEGFP3-ADH1t | This study |
| S. cerevisiae strains | ||
| CEN.PK2-1C | MATa ura3-52 trp1-289 leu2-3,112 his3Δ 1 MAL2-8C SUC2 | EUROSCARF |
| TMB4114 | CEN.PK2-1C trp1::TRP1 leu2::LEU2 his3− ura3− | This study |
| TMB4112 | CEN.PK2-1C gpd1::TRP1 gpd2::LEU2 his3− ura3− | This study |
| TMB4122 | CEN.PK2-1C YEpJK01 trp1::TRP1 gpd2::LEU2 his3::HIS3 | [26] |
| TMB4140 | CEN.PK2-1C trp1::TRP1 leu2::LEU2 his3::YIpJK01 ura3::YIplac211 | [26] |
| TMB4144 | CEN.PK2-1C gpd1::TRP1 gpd2::LEU2 his3::YIpJK01 ura3::YIplac211 | [26] |
| TMB4145 | TMB4114 his3::HIS3 YEpJK03 | This study |
| TMB4146 | TMB4112 his3::HIS3 YEpJK03 | This study |
2.2. Molecular biology methods
Plasmid DNA was prepared using the GeneJET Plasmid Miniprep Kit (Thermo Scientific, USA). Restriction and modification enzymes as well as T4 DNA ligase were obtained from the same manufacturer. The QIAquick gel extraction kit (QIagen, Hilden, Germany) was used for DNA extractions from agarose. All nucleotides were ordered at Eurofins (Germany). All genetic constructs were checked by sequencing (Eurofins, Germany). The primers used in the study are listed in Table 2.
Table 2.
Primers used in the present study.
| Name | Description | Sequence |
|---|---|---|
| SpeI_yEGFP3_f | Forward primer for amplification of yEGFP3 from pyEGFP3 | GAACTAGTAAAAATGTCTAAAGGTGAAG |
| XhoI_yEGFP3_r | Reverse primer for amplification of yEGFP3 from pyEGFP3 | GACTCGAGGTGTTATTTGTACAATTCATCCATAC |
Competent cells of E. coli NEB 5α were prepared and transformed by the method of [22] and yeast transformations were performed using the lithium acetate method [18]. E. coli transformants were selected on lysogeny broth (LB) plates [43] with 50 μg/ml ampicillin (IBI Shelton Scientific, Shelton, USA). S. cerevisiae transformants were selected on YNB.
2.3. Plasmid construction
Plasmid YEpJK03 was constructed by ligation of two DNA fragments using T4 DNA ligase: (1) The yEGFP3 fragment that was PCR amplified from pYGFP3, using the primers SpeI_yEGFP3_f and XhoI_yEGFP3_r, digested with SpeI and XhoI and finally purified, and (2) 426GPD digested with SpeI and XhoI. The ligation mix was then used to transform E. coli.
2.4. Construction of yeast strains
TMB4112, TMB4114, TMB4145, TMB4146 (Table 1) were all constructed using CEN.PK2-1C as parental strain. The strains were made prototrophic by sequentially transforming the CEN.PK2-1C strain with (i) the required expression cassettes for LEU2, TRP1 or HIS3, (ii) the GPD1 and GPD2 deletion cassette and (iii) YEpJK03 plasmid.
TMB4114 and TMB4112 were constructed by transformation of CEN.PK2-1C with TRP1, LEU2 and HIS3 expression cassettes or the GPD1 (TRP1) and GPD2 (LEU2) deletions cassettes as described in Knudsen et al. [26]. TMB4145 and TMB4146 were constructed by transforming TMB4114 or TMB4112 with YEpJK03. Double deletions were verified as described in [26].
2.5. Shake flask cultivations
Cells were taken from YNB plates and inoculated in 50-ml falcon tubes containing 5 ml defined medium buffered to pH 6.5 and complemented with 2% glucose [52], and incubated in a rotary shake incubator set to 180 rpm and 30 °C. After 16–20 h, cells from the pre-culture were inoculated with an optical density (OD620nm) of 0.1–0.2 in 0.5 l baffled shake flasks containing 50 ml of the same media. Shake flask cultivations were performed in two separate experiments for the wild type strain and once for the deletion strain.
2.6. Oxygen-limited batch cultivations in stirred tank reactor (STR) without air sparging
Pre-cultures were grown in 0.5 l baffled shake flasks containing 50 ml defined media buffered to pH 6.5 and complemented with 2% glucose [52]. After approximately 10 h, the cells were transferred to Multifors HT bioreactors (Infors HT, Switzerland) at an OD of 0.01–0.02 and a working volume of 1 l. The pH was maintained at 5.5 with 2 M NaOH and the temperature kept at 30 °C. No aeration was applied, and the stirring was kept at 400 rpm. The pH electrodes (Mettler Toledo, OH, USA) were calibrated using a two point calibration method (pH 4.0 and 7.0) according to standard procedures provided by the manufacturer. Batch cultivations were performed in two separate experiments.
2.7. Chemostat cultivations
Pre-cultures were cultivated for 8–12 h in baffled shake flasks as described above. The shake flask grown cells were then used to inoculate 500–750 ml working volume in Sartorius bioreactors (Sartorius Stedim Biotech, German) to an OD of 0.01–0.02. The pH was maintained at 5.5 with 2 M NaOH, aeration was set to 2.5 vvm, the stirring was kept constant at 800 rpm and the temperature was kept at 30 °C. The pH and dissolved oxygen tension (DOT) electrodes (Mettler Toledo, OH, USA) were calibrated according to standard procedures provided by the manufacturer using two point calibration (pH 4.0 and 7.0, gassing with oxygen (100%) and nitrogen (0%), respectively). Growth was followed by measuring the CO2 content in the exhaustion gas. The composition of the outgoing gas from batch cultivations was monitored by a mass spectrometer (Prima Pro Process MS, Thermo Fisher Scientific, Winsford, UK). Once the glucose was depleted a feed corresponding to a dilution rate of D = 0.05 h−1 was initiated. Steady state was assumed after 5 retention times and stable CO2 levels. Samples for HPLC, OD, dry weight and flow cytometry were taken at one retention time intervals. Three sample points were taken at each steady state. For each dilution rate, the population was allowed to reach a new steady state (monitored by steady CO2 levels) for at least 3 retention times before taking samples.
2.8. Accelerostat cultivations
For accelerostat cultivation [38], cell populations were first allowed to reach steady state at a dilution rate of 0.05 h−1 in aerobic, glucose-limited chemostat cultures (see above). The dilution rate was then stepwise increased with an acceleration factor of ≈0.01 h−1, using a calibrated peristaltic pump manually adjusted every hour. Samples for HPLC, OD and flow cytometry were taken at each dilution rate. Accelerostat cultivations were performed in two separate experiments.
2.9. Analyses
Samples for measurement of optical density (OD) and flow cytometry analysis of the batch cultivations were analysed directly while samples for HPLC were kept at −20 °C until analysis. Growth was monitored by measuring OD at 620 nm with a Shimadzu UV mini 1240 spectrophotometer (Shimidzu, Kyoto, Japan) or a Ultraspec 2100 pro spectrophotometer (GE Healthcare Life sciences, Sweden). Cell dry weight was determined by filtering 5 ml cultivation broth in pre-weighed 0.45 μm pore size membrane filter (Pall Corporation, Port Washington, NY, USA). Filters were washed with distilled water and dried at 350 W in a microwave oven. In case when only OD620 was determined, the DW (g/l) was calculated from OD according to an empirical formula describing the correlation for CEN.PK2-1C strain, DW = 0.17 × OD + 0.17. The concentrations of glucose, acetate, ethanol, glycerol, and pyruvate were determined by HPLC (Agilent 1100, Agilent Technologies, CA, USA) with a 300 mm × 7.8 mm Aminex HPX-87H ion exchange column (Bio-Rad, Hercules, CA, USA), refractive index detector (RID Agilent 1200, Agilent Technologies, CA, USA) and UV detector (Agilent 1100, Agilent Technologies, CA, USA) set to 210 nm. The mobile phase was 5 mM H2SO4(aq.), temperature 60 °C and flow rate 0.6 ml/min. A BD Accuri™ C6 (Becton-Dickinson, NJ, USA) flow cytometer equipped with a CSampler was used for the flow cytometric analysis of the shake flasks cultivations and oxygen limited batch cultivations. A blue laser (λ = 488 nm) was used for GFP excitation and fluorescence emission levels were measured using a band pass filter at 533/30 nm. Light scattering and florescence emission were standardised using Spherotech 8-Peak validation beads according to the manufactures instructions. Cell viability was determined by flow cytometry analysis, using the BD Accuri™ C6 flow cytometer, of cells stained with propidium iodide (PI) as described previously [7]. A BD FACSAria III (Becton-Dickinson, NJ, USA) flow cytometer was used for the single-cell analysis of the accelerostat experiments and continuous cultures. A blue laser (λ = 488 nm) was used for GFP excitation and fluorescence emission levels were measured using a band pass filter at 529/28 nm. Samples for flow cytometry were centrifuged for 1 min at 3000 × g and 4 °C, and resuspended in 0.9% saline solution. For the flow cytometric analysis of the continuous cultivations, samples were frozen in phosphate buffer, pH 7.0, 25% glycerol at −80 °C before being thawed and analysed. 10,000 events were recorded with a rate of approximately 1000 events per second. The flow cytometry raw data was exported as fcs files by the flow cytometer FACSAria III and Accuri C6, then imported into MATLAB and processed and analysed using MatLab®R2010b (The MathWorks, Inc., Natick, MA, USA).
3. Results
3.1. GPD2 promoter activity in glycerol-negative and glycerol-positive strains during aerobic batch cultivation in shake flasks
The GPD2 promoter activity was investigated in reporter strains carrying the GFP reporter controlled by the GPD2 promoter, as has been described previously [26]. The reporter construct was placed in strains with or without double deletion of GPD1 and GPD2, i.e. in strains with different ability to re-generate NADH, which earlier has been shown to directly influence regulation of GPD2 transcription [26]. The cultivations were performed in aerobic shake flasks with an excess of glucose (20 g/l) leading to overflow metabolism in the glucose growth phase and the need for cytosolic NADH oxidation via the glycerol production pathway.
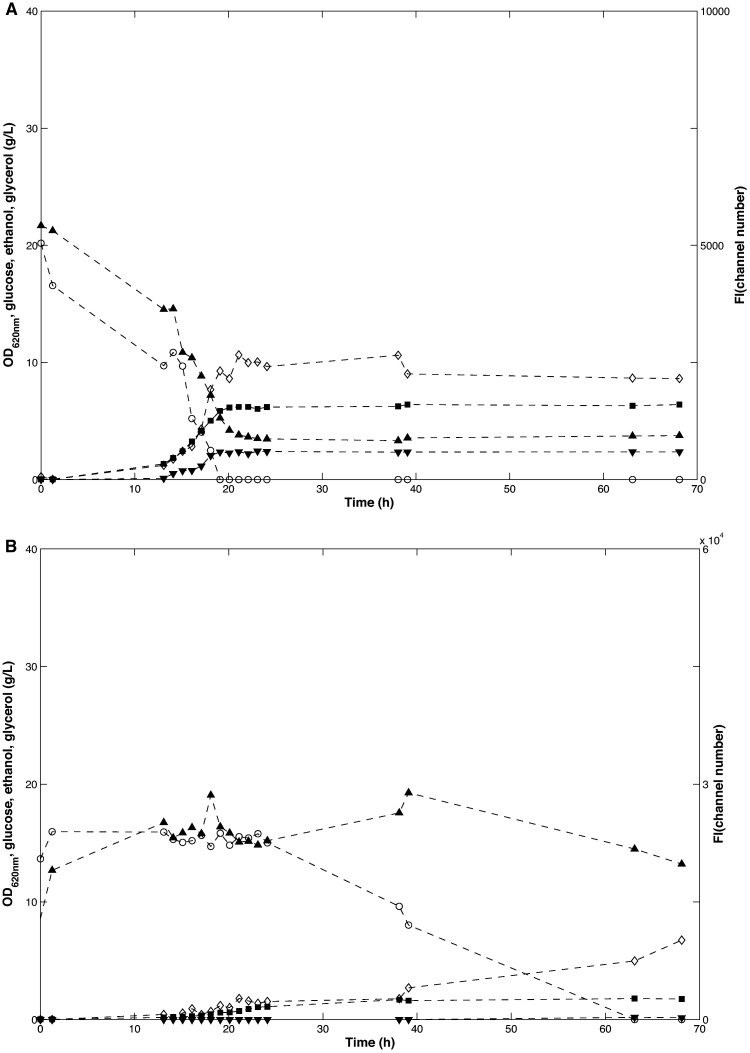
For the wild type reporter strain TMB4140, four typical growth stages [13] of aerobic batch cultivation mode were observed (Fig. 1A), i.e. respiro-fermentative growth on glucose (0–24 h), diauxic shift (∼24 h), respiratory growth on ethanol (24–40 h) and finally stationary phase (>40 h). The mean fluorescence intensity (FI) of the population had an initial value of 5410 ± 10 channel numbers (ch.nr.) and decreased steadily to 1620 ± 140 ch.nr. until around 24 h, which coincided with glucose depletion (Fig. 1A). The maximum glycerol production rate (0.05 ± 0.00 g/gDW/h) occurred at the beginning of the batch cultivation, which was also mirrored by the higher arithmetic mean GFP level and thus the highest mean GPD2p activity.
Fig. 1.
Batch cultivations in aerobic shake flasks. Glucose (g/l), extracellular metabolites (g/l), OD620nm and FI* were recorded during cultivations in shake flasks. A TMB4140 (WT) vertical lines indicate end of lag growth phase (≈15 h, defined as start of decreased glucose concentration) end of exponential phase (≈24 h, defined as start of decreased ethanol concentration) and entry to stationary phase (≈40 h, defined as the time point where after there was no further increase in OD). B TMB4144 (gpd1Δgpd2Δ) vertical lines indicate end of lag phase (≈22 h defined as start of decreased glucose concentration) and entry to stationary phase (≈60 h, defined as time point where after there was no further increase in OD). Legends: Glucose (circle), Glycerol (downwards triangle), Ethanol (diamond), OD (square), FI (upwards triangle). The experiments were performed in biological duplicates. One representative experiment is shown.
*FI is the fluorescence intensity per cell, and was measured with BD Accuri™ C6 (Becton-Dickinson, NJ, USA) equipped with a 488 nm laser for excitation of GFP, and a band pass filter of 533/30 nm for collection of the emission.
The gpd1Δgpd2Δ reporter strain (TMB4144) was cultivated under the same conditions as the wild type strain (TMB4140) but displayed a different growth profile (Fig. 1B). It had a long lag phase (∼20 h vs. 15 h), approximately 2-fold lower growth rate (μmax of 0.15 ± 0.01 h−1 as compared to 0.29 ± 0.00 h−1) and as a consequence a delayed stationary phase (60 h vs. 40 h). Furthermore, no glycerol accumulation was observed and the formation of ethanol was 1.25 fold higher (0.45 ± 0.08 g/gDW/h vs.0.36 ± 0.0 g/gDW/) and the maximal OD was lower (OD620 = 26 ± 0.0 as compared to 31 ± 0.3). The mean FI was also significantly higher in the gpd1Δgpd2Δ strain, which is in accordance with a previously performed study [26]. The FI increased steadily from an initial value of 12300 ± 600 ch.nr. until it reached a maximum of 45500 ± 5500 ch.nr. (8 times the highest value measured for TMB4140 and approximately 39-fold higher than yeast auto-fluorescence) at the same time as glucose consumption started and exponential growth was first observed. After this point, the mean FI decreased to a steady level of around 2500 ch.nr. at around 65 h when both glucose and ethanol were totally consumed. At this phase of the cultivation the FI of the reporter strain was equal to the autofluorescence of a control yeast strain without the GFP gene. It is worth noting that the FI signal increased almost linearly for the gpd1Δgpd2Δ strain during the lag phase when consumption of glucose was unmeasurable and there was no increase in cell concentration.
3.2. GPD2 promoter activity during fermentative growth in STR without sparging
In order to further investigate the response of the reporter strains under conditions where the GPD2p is known to be induced, batch cultivations were performed in stirred tank reactors (STRs) with glucose as the sole carbon source but without air sparging. The non-sparged setup made it possible to monitor the transition from an aerobic environment at the time of inoculation to an oxygen-limited environment during growth on glucose, which causes a rapid depletion of the remaining oxygen. Under these conditions, the TMB4140 (wild type) strain reached a lower final OD than under aerobic conditions, OD 6.6 ± 0.2 vs OD 31 ± 0.3 (Figs. 2A and 1A). However, the maximum growth rate on glucose was the same in STR cultivations without sparging as in the case of the aerobic shake flask, i.e. 0.3 ± 0.0 h−1. The concentration of extracellular metabolites increased until glucose was exhausted and growth ceased, which in combination with the lack of respiratory assimilation of ethanol demonstrated the lack of oxygen in the STR.
Fig. 2.
Batch cultivations in aerobic stirred tank reactors (STRs) without air sparging. Glucose (g/l), extracellular metabolites (g/l), OD620nm and FI* were recorded during cultivations in stirred tank reactors. A TMB4140 (WT) and B TMB4144 (gpd1Δgpd2Δ). Legends: Glucose (circle), Glycerol (downwards triangle), Ethanol (diamond), OD (square), FI (upwards triangle). The experiments were performed in biological duplicates. One representative experiment is shown.
*FI is the fluorescence intensity per cell, and was measured with BD Accuri™ C6 (Becton-Dickinson, NJ, USA) equipped with a 488 nm laser for excitation of GFP, and a band pass filter of 533/30 nm for collection of the emission.
For the wild type reporter strain, a similar dynamic profile of GPD2 promoter activity was observed during oxygen limited batch cultivations as during cultivation in the aerobic shake flasks, i.e. the FI signal was highest at the start of cultivation (5000 ± 400 ch.nr.). Thereafter, the FI signal decreased during the next 20 h to reach yeast autofluorescence level (1200 ± 70) at the time point when glucose was depleted, thus demonstrating that the GPD2 promoter was inactive. From this point of the cultivation the bioreactor was under semi-anaerobic conditions and the FI signal, ethanol, glycerol, and acetate concentrations remained constant until the cultivation was terminated (Fig. 2A). GFP has previously been reported to require oxygen to form a functional fluorescent fluorochrome [23] and it may be hypothesized that the lack of GFP signal was due to lack of dissolved oxygen in the STR. However, the oxygen supplied from the stirring in the non-sparged STR or during sampling was enough for measuring GFP fluorescence in the gpd1Δgpd2Δ reporter strain. It has previously been shown that the average maturation time of GFP in E.coli is around 6 min [21], which allows for maturation of the protein during the sample handling.
The gpd1Δgpd2Δ (TMB4144) cells did not produce glycerol and were thus unable to re-oxidize NADH generated from anabolic reactions, which caused a lower growth rate (μmax of 0.16 ± 0.01 h−1) and a 3-fold lower final cell density (OD < 2.0 ± 0.2) compared to aerobic conditions during shake flask cultivations (Figs. 1B and 2B). Furthermore, complete glucose consumption was not reached and the ethanol levels were 23% lower than for the wild type after 70 h. The cellular FI was significantly higher than for the wild type population throughout the whole fermentation and never fell below 15000 ch.nr. Unlike the wild type strain the FI signal of the gpd1Δgpd2Δ mutant increased from time point zero to a relatively constant level of 25000 ch.nr. before finally declining at the end of the fermentation.
3.3. GPD2 promoter activity at different growth rates in accelerostat mode
To investigate potential correlations between GPD2 promoter activity and growth rate, a set of fully aerobic accelerostat cultivations were performed, starting from chemostat mode at D = 0.05 h−1 with a stable linear increase of ΔD ≈ 0.01 h−1 to a final D = 0.40 h−1. The accelerostat is in general valued as a rapid and effective way of screening microbial behaviour at a range of different dilution rates and has previously been applied to determine correlations between growth-rate and mode of sugar metabolism in budding yeast [50]. Additionally the accelerostat set-up allowed for a rapid investigation of the transition from low growth rates and fully respiratory metabolism to higher growth rates where overflow metabolism occurs (i.e. at the critical dilution rate where ethanol formation is observed, Dethanol), and also a second transition to washout (Dwashout), where the cells are proliferating at maximum growth rate. In order to ensure complete respiratory metabolism at low dilution rates, the aeration and stirring rate were set high, i.e. 2.5 vvm and a stirring rate of 800 rpm. Consequently, the respiratory quotient (RQ) value was approximately 1 at dilutions rates below 0.3 h−1, demonstrating non-limiting oxygen levels in the STRs (Fig. 3E and F).
Fig. 3.
Accelerostat cultivations in highly aerobic stirred tank reactors (STR). Glucose (g/l), extracellular metabolites (g/l), OD620nm and FI* were recorded during accelerostat experiments. A, C, E TMB4140 (WT) and B, D, F TMB4144 (gpd1Δgpd2Δ). Legends: A, B Glucose (circle), Ethanol (diamond) and Glycerol (cross). C, D FI (cross), OD (triangle). E, F Respiratory quotient (RQ). The experiments were performed in biological duplicates. One representative experiment is shown.
*FI is the fluorescence intensity per cell, and was measured with BD FACSAria III (Becton-Dickinson, NJ, USA) equipped with a 488 nm laser for excitation of GFP, and a band pass filter of 533/30 nm for collection of the emission.
When comparing the metabolic responses of the two strains in relation to increases in dilution rate during the accelerostat cultivation, the gpd1Δgpd2Δ (TMB4144) strain produced ethanol at a slightly lower dilution rate (Dethanol) than the wild type strain (TMB4140), 0.20 h−1 and 0.25 h−1, respectively (Table 3 and Fig. 3A and B). Washout from the bioreactor was for both strains observed at a dilution rate of Dwashout = 0.4 h−1, which was evident from the declining CO2 exhaustion (data not shown), biomass and ethanol concentrations and the increasing glucose concentration observed at D > 0.40 h−1. The strains performed notably different in terms of glycerol production. No production was observed for the gpd1Δgpd2Δ mutant strain, whereas the wild type strain produced a measurable amount of glycerol when at D = 0.35 h−1 or higher.
Table 3.
Critical dilution rates of the accelerostat experiment and chemostat experiment.
| Strain |
Dgly (h−1) |
Dethanol (h−1) |
Dwashout (h−1) |
Dgly (h−1) |
Dethanol (h−1) |
|---|---|---|---|---|---|
| Accelerostat | Chemostat | ||||
| TMB4140 | 0.35 | 0.25 | 0.4 | – | 0.2 |
| TMB4144 | – | 0.2 | 0.4 | – | 0.25 |
With regards to GPD2 promoter regulation, the activity indeed correlated to the dilution rate for both strains, but was at the same time strain-dependent. The mean FI level per cell was 20–30 % higher for the gpd1Δgpd2Δ mutant at dilution rates below 0.30 h−1 and above 0.40 h−1 (Fig. 3C and D). However, between D = 0.30 and 0.40 h−1, both strains produced similar FI signals, which indicates that there was no need for de novo synthesis of the Gpd2p enzyme at this point under the applied high-aeration conditions. The FI signal did not increase with the onset of the Crabtree effect i.e. the onset of ethanol formation and increase of the respiratory quotient. Instead, an increase in FI signal was first observed in the gpd1Δgpd2Δ mutant at a dilution rate close to where glycerol started to be measurable for the wild type, i.e. D = 0.40 h−1. As can be seen from Fig. 3, the channel number values of FI was lower than for the above mentioned batch cultivation experiments (Fig. 1, Fig. 2), which is due to that the raw data were collected with different FC instruments.
3.4. GPD2 promoter activity at different growth rates in chemostat mode
The accelerostat experiment gave insight about the need for glycerol synthesis and GPD2 promoter activity at different growth rates under dynamic conditions. To further elucidate the activity of the GPD2 promoter at different specific growth rates and in proper steady-state environments, a number of glucose-limited chemostat cultivations were performed. In glucose-limited chemostat cultures the glucose concentration, the specific growth rate, and therefore the rate of glycolysis can be controlled. In this way, glucose catabolite repression of respiratory enzymes and alcoholic fermentation can be controlled as needed. Two fully respiratory growth rates were chosen i.e. D = 0.05 and 0.10 h−1, two intermediate growth rates D = 0.20 and 0.25 h−1 and one high respiratory–fermentative i.e. D = 0.30 h−1. The same trend as in the accelerostat experiment was observed. The FI signal was high at low dilution rates and decreased with increasing dilution rate before reaching a plateau (Fig. 4). The FI signal of the mutant strain was higher than that of the wild type, but both reached the same bottom plateau, which was at yeast autofluorescence level, indicating a full inactivation of the GPD2 promoter at D = 0.30 h−1. The glycerol production seen for the wild type and the observed increase in FI for the gpd1Δgpd2Δ mutant strain in the accelerostats, were not detected in the chemostat cultivations. This was also expected since the highest dilution rate was set below the dilution rate where glycerol was observed in the accelerostat cultivation (<Dglycerol = 0.35 h−1).
Fig. 4.
Chemostat cultivations in highly aerobic stirred tank reactors (STR). Glucose (g/l), extracellular metabolites (g/l), OD620nm and FI* were recorded during chemostat experiments. A, C, E TMB4140 (WT) and B, D, F TMB4144 (gpd1Δgpd2Δ). Legends: A, B Glucose (circle), Ethanol (diamond) and Glycerol (cross). C, D FI (cross), OD (triangle). E, F Respiratory quotient (RQ). The experiments were performed in biological duplicates.
*FI is the fluorescence intensity per cell, and was measured with BD FACSAria III (Becton-Dickinson, NJ, USA) equipped with a 488 nm laser for excitation of GFP, and a band pass filter of 533/30 nm for collection of the emission.
The observed Dethanol was lower for the gpd1Δgpd2Δ mutant (Dethanol = 0.20 h−1) than for the wild type strain (Dethanol = 0.25 h−1), which was also in agreement with the accelerostat experiment (Table 3). There was no accumulation of metabolites in either of the two strains, indicating a fully respiratory metabolism at dilution rates between 0.05 and 0.20 h−1. Biomass yield for the wild-type strain was 0.47 ± 0.09 g/g below D = 0.25 h−1, which is in accordance with previously performed glucose-limited aerobic chemostats where the biomass yield over glucose has been reported to be around 0.5 g/g below a dilution rate of 0.23 h−1 [35]. For the gpd1Δgpd2Δ mutant, the biomass yield below Dethanol = 0.20 h−1 was slightly higher (0.58 ± 0.04 g/g). However, at D = 0.3 h−1 which is well above Dethanol for both strains, the biomass yield was 20% lower for the gpd1Δgpd2Δ strain compared to the wild type. Furthermore, the ethanol yield was 40 % higher for the gpd1Δgpd2Δ mutant compared to the wild type strain. This is in accordance with a previously performed comparison of the performance of glycerol-negative and wild type strains in aerobic batch cultivations [31].
The mean FI signal decreased with increasing dilution rate in a similar manner as the accelerostat experiments for both strains, with the gpd1Δgpd2Δ strain giving the higher signal. In accordance with the accelerostat experiments the FI signals decreased with increasing dilution rate until the onset of the Crabtree effect. However, in contrast to the accelerostat experiments the chemostat FI signals remained constant at dilution rates above the onset (>Dethanol).
3.5. Evaluation of heterogeneity of GPD2 promoter activity within the population
Single-cell analysis of the FI signal from the reporter strains can reveal if sub-populations with different regulation of the GPD2 promoter are present, but this was not observed in any cultivation performed within the study. However, a subpopulation with low FI appeared at the end of the oxygen-limited gpd1Δgpd2Δ cultivation (>60 h) (Fig. 5). The subpopulation also stained positive with propidium iodide (PI) (data not shown), indicating that it had acquired a permeabilized membrane phenotype [7].
Fig. 5.
Population heterogeneity in cellular FI* during batch cultivations in shake flasks and stirred tank reactors (STR) without air sparging figure A, and B respectively. Sample points: 1.25 h, 13.1 h, 19.1 h, 21.1 h and 63.1 h. Both strains TMB4140 (WT) and TMB4144 (Δgpd1Δgpd2) are shown. The line shows the arithmetic mean of the autofluorescence of yeast cells grown over night in shake flasks.
*FI is the fluorescence intensity per cell, and was measured with BD Accuri™ C6 (Becton-Dickinson, NJ, USA) equipped with a 488 nm laser for excitation of GFP, and a band pass filter of 533/30 nm for collection of the emission.
Cell populations grown in aerobic shake flasks had a positive correlation between FI level and FI standard deviation. An often applied measure for population heterogeneity is the coefficient of variation (CV), which is defined as the standard deviation divided by the arithmetic mean value (standard deviation/mean value). After normalizing the standard deviation to the FI mean, which is analogous to how the CV value is calculated, there were no longer any apparent differences in cell-to-cell variation of GPD2p activity between strains and cultivation conditions. The similar degree of population heterogeneity of the two strains during different phases of shake flask cultivations and oxygen availability is also apparent when comparing broadness of the peaks in the FI histograms (Fig. 5).
From the accelerostat and chemostat cultivations no correlation between population heterogeneity and dilution rate was observed for any of the reporter strains (Fig. 6). However, while cell populations that proliferated in accelerostat mode displayed no obvious difference in cell-to-cell variation between the two strains, the CV values obtained from continuous chemostat cultures showed a difference between the wild type and Δgpd1Δgpd2 strain (CV = 0.25–0.37 compared to CV = 0.23–0.30 at D = 0.05–0.28 h−1, respectively). Nevertheless, distinct subpopulation with different degree of FI intensity could not be observed in any of the cultivations, and the variation in GPD2 promoter activity between cells is probably therefore not a consequence of programmed differential gene regulation other than stochastic differences in activity.
Fig. 6.
Population heterogeneity in cellular FI* during accelerostat and chemostat cultivations. Figures A and B respectively. Samples are shown from dilution rates of 0.4, 0.3, 0.2 and 0.06 h−1 (accelerostat) and 0.32, 0.22 and 0.06 h−1 (chemostat). The line shows the arithmetic mean of the autofluorescence of yeast cells grown over night in shake flasks.
*FI is the fluorescence intensity per cell, and was measured with BD FACSAria III (Becton-Dickinson, NJ, USA) equipped with a 488 nm laser for excitation of GFP, and a band pass filter of 533/30 nm for collection of the emission.
3.6. Comparison of GPD2 and TDH3 promoters for controlling GFP expression from multicopy plasmid
In order to further assess the value of the GPD2 promoter for recombinant protein production, it was placed on a 2 μ multicopy plasmid (yeast episomal plasmid, YEp) and a series of oxygen-limited cultivations in non-sparged glass vials were performed. The mean FI obtained for the reporter strain was as expected significantly higher in the gpd1Δgpd2Δ strain than the wild type strain, irrespective of if the GPD2p-GFP construct was integrated (20-fold higher) or placed on a multicopy plasmid (21-fold higher). However, both the mean FI level that was reached and the heterogeneity of the population were significantly higher with the GPD2p-GFP construct on a multicopy plasmid than on a yeast integrative plasmid (YIp) (Fig. 7). The observed difference between the YEp and YIp systems has been described previously [26], and can be speculated to be caused by higher gene copy number as well as larger variation in plasmid replication kinetics.
Fig. 7.

Comparison between FI*, when GFP expression is under the control of TDH3 and GPD2 promoters. Notice the logarithmic y-axis. Reporter constructs were placed on multicopy plasmids and transformed into both wild type and gpd1Δgpd2Δ strains.
*FI is the fluorescence intensity per cell, and was measured with BD Accuri™ C6 (Becton-Dickinson, NJ, USA) equipped with a 488 nm laser for excitation of GFP, and a band pass filter of 533/30 nm for collection of the emission.
For comparison, a reporter strain expressing the GFP gene under the control of the glyceraldehyde-3-phosphate dehydrogenase (TDH3) promoter [5], [30] was also constructed and characterised in parallel. Surprisingly, the mean FI obtained with the TDH3 promoter was 2-fold higher when placed in the gpd1Δgpd2Δ strain (TMB4146) as compared to when it was placed in the wild type strain (TMB4145). Nevertheless, the mean FI obtained for the gpd1Δgpd2Δ reporter strain expressing GFP from the GPD2 promoter was 2-fold higher than the one obtained from the TDH3 promoter (Fig. 7).
4. Discussion
A window of operation where glycerol-negative yeast can be cultivated nearly as efficiently as wild type yeast was identified by flow cytometric analysis of reporter strains expressing GFP under the control of the GPD2 promoter. Cells growing at D = 0.3 h−1 with fully respiratory metabolism were shown to have an inactivated GPD2 promoter regardless of strain capacity for GPD-catalysed NADH oxidation. With this follows that there was no need for Gpd2p activity under the applied process conditions and that glycerol-negative yeast can be produced in an efficient way as long as aeration is sufficiently supplied. GPD deletion strains have previously been reported to grow significantly slower than wild type yeast even under aerobic conditions [3], [6]. However, a significant difference to these studies was that the GPD deletion strain here was grown with high aeration (2.5 vvm), which led to an RQ-value of 1, whereas the previous experiments have been performed in either shake flasks or at a lower aeration rate (0.3 vvm) [27], most likely resulting in oxygen-limited conditions. It has previously been shown that glycerol production can be regulated by controlling oxygen uptake rate [56]. Weusthuis et al. demonstrated that above a critical threshold value of oxygen transfer rate, glycerol production was not observed, which was explained by that cells were able to close their redox balance through respiration.
The early phase of batch cultivations were characterised by glucose rich conditions as well as a short lag phase. For the wild type reporter strain a correlation was observed between GPD2p activity and growth phase during both aerobic and oxygen-limited batch. This may be due to different NADH/NAD+-ratios at different phases of cultivation and thus at different growth rates, which has previously been indicated as determinant for cellular redox state [37]. Indeed, a negative relationship between cellular GPD2 promoter activity and growth rate was clearly visible for both the chemostat and the accelerostat continuous cultivations. Cells grown at dilution rates below the critical dilution rate where fermentative metabolism occurs (Dethanol < 0.2) displayed the highest GPD2p activity. The same relationship, i.e. high GPD2 promoter activity at low D, has previously been measured in a transcriptomic study of yeast grown at different dilution rates in glucose-limited aerobic chemostats [40]. From the transcriptome data generated by Regenberg et al. [40], it can be read that both the GPD2 promoter and the mitochondrial glycerol-3-phosphate dehydrogenase (GUT2) promoter are up-regulated at low dilution rates, whereas the mitochondrial external NADH dehydrogenase (NDE1) promoter is inactive. At higher dilution rates the inverse relationship was measured, which suggests that Gpd2p is involved in the glycerol-3-phosphate (G3P) shuttle at low dilution rates and fully respiratory metabolism, but not at intermediate dilution rates below the value where there is a need for glycerol synthesis.
As particularly observed for the gpd1Δgpd2Δ strain, there was a clear difference if the cells were grown in aerobic shake flasks or in a STR without aeration, which reflects the responsiveness of the GPD2 promoter to oxygen levels, or perhaps more stringently to the need for non-mitochondrial oxidation of NADH generated from anabolic reactions. During the aerobic batch cultivations the FI per cell increased while no glucose consumption and no increase in cell concentration were observed. As the exponential growth phase started, a decrease in the FI per cell was observed until glucose was depleted and the cells started to assimilate ethanol by respiration. At this stage of batch cultivation, the FI intensity was the highest observed throughout the study, and it was even higher than the well-known TDH3 promoter. After this point the GPD2 promoter activity was down-regulated and FI levels decreased as a consequence of GFP degradation and due to growth-related dilution effect [42]. At its highest level, the GPD2 promoter activity was 20-fold higher in the glycerol-deficient mutant than the wild type strain under these non-sparged conditions. The fact that FI reached a maximum just before growth initiated implies that the activity of Gpd2p is crucial to maintain NADH balance during the initial protein biosynthesis phase to adapt the cell proteome to its new environment, and before the start of cell proliferation can occur. This hypothesis is compatible with earlier observations by Valadi et al. [48], who found that a gpd2Δ strain had a significantly longer lag phase during anaerobic growth than a wild type strain.
It should also be highlighted that the high activity of the GPD2 promoter in the gpd1Δgpd2Δ strain at oxygen-limited conditions, together with its nearly complete inactivation at high aeration and intermediate dilution rates, can be used as an inducible system for production of recombinant proteins. Heterologous gene expression controlled by the GPD2 promoter does thus not depend on keeping low glucose levels, which is the case for the galactose-inducible (GAL) promoters [28], nor is it dependent on the addition of inducer compounds such as doxycycline [4].
From a process perspective it may be advantageous to separate the production process into two steps:
-
i.
An aerobic fed-batch cultivation step operated just below Dethanol to generate biomass without diversion of carbon to by-products and where the GPD2 promoter is inactive;
-
ii.
An anaerobic or oxygen-limited phase where the GPD2 promoter is hyperactive.
Control of the GPD2 promoter, and thus control of the production of recombinant protein can be performed by first simply decreasing the aeration rate and then adjusting the media inlet feed rate to allow for maximum growth rate.
However, the exact procedures for optimal cell growth and subsequent GPD2p-induced bio-production of proteins or chemicals needs to be properly addressed in further optimisation studies for each specific case. A separation of the cell cultivation phase from the production phase may be important if the specific protein or specific metabolic pathway that should be induced is toxic or inhibitory to cell growth. The proposed induction strategy may thus in some cases lead to higher space time yield than a traditional non-induced system with constitutive expression. The use of a gpd1Δgpd2Δ strain as production strain background may yet be problematic due to its relatively low robustness to adverse process conditions such as high osmotic pressure from high glucose levels. The glycerol-deficient yeast has however recently been adapted by evolutionary engineering to withstand high osmotic pressure with similar growth phenotype as wild type yeast [19].
Finally, glycerol-deficient yeast is a promising platform for NADH-dependent whole-cell biocatalytic reactions, for example for conversion of carbonyl compounds to high-value alcohols used as intermediates for pharmaceutical synthesis [57]. However, upstream production of the whole-cell biocatalyst has previously been considered to be a hinder in its implementation in biotransformation applications. The results presented herein demonstrate that glycerol-deficient yeast can be cultivated efficiently and illustrates that up-stream production can be made efficient if substrate feed rate is controlled and aeration of the bioreactor can be kept at high level. The potential trade-off between efficient upstream production of NADH-accumulating yeast and efficient whole-cell biocatalysis can be diminished. The cells can be cultivated efficiently in a first stage followed by a second biocatalytic stage with conditions adapted to drive NADH-dependent reductions. Furthermore, under the same environmental conditions that result in high availability of cytosolic NADH, the GPD2 promoter is also hyperactive and could be therefore be used to reach high enzyme levels. Controlling the presence of a specific enzyme in a multi-enzyme biocatalytic cascade can also be used to separate the time a specific reaction step should occur.
5. Conclusions
Process conditions were identified where the GPD2 promoter was either completely inactive on one hand, or hyperactive on the other. By placing the expression vector in a gpd1Δgpd2Δ strain background the strength of the GPD2 promoter benchmarks with the strong constitutive TDH3 promoter, which demonstrates its potential for recombinant gene expression. Furthermore, the inducible character of the promoter provides a means of separating production and cultivation phases by simply regulating dissolved oxygen level and substrate feed rate. Possible scenarios where this may be desirable include usage for the control of toxic protein production, or for the control of metabolic pathways in which intermediates or even the product negatively influence cell proliferation efficiency.
Conflict of interest
The authors declare that they have no competing interest.
Author contribution
JDK and MC contributed equally in the design of the shake flask and STR experiments. JDK,TJ, AEL and MC contributed equally in the design of the accelerostat and chemostat experiments. JDK performed the experimental work and wrote the manuscript together with MC. TJ and AEL helped in drafting the manuscript. All authors have read and approved the submission of the manuscript.
Acknowledgements
This work was financially supported by the Swedish Research Council (Vetenskapsrådet) and The Swedish Research Council FORMAS. Stefan Morberg is acknowledged for technical assistance with the flow cytometry analysis
Footnotes
Available online 15 June 2015
Contributor Information
Jan Dines Knudsen, Email: jan.knudsen@tmb.lth.se.
Ted Johanson, Email: ted.johanson77@gmail.com.
Anna Eliasson Lantz, Email: aela@kt.dtu.dk.
Magnus Carlquist, Email: magnus.carlquist@tmb.lth.se.
References
- 1.Abbott D.A., Zelle R.M., Pronk J.T., van Maris A.J. Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: current status and challenges. FEMS Yeast Res. 2009;9:1123–1136. doi: 10.1111/j.1567-1364.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- 2.Albertyn J., Hohmann S., Thevelein J.M., Prior B.A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cellular Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansell R., Granath K., Hohmann S., Thevelein J.M., Adler L. The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J. 1997;16:2179–2187. doi: 10.1093/emboj/16.9.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belli G., Gari E., Piedrafita L., Aldea M., Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bitter G.A., Egan K.M. Expression of heterologous genes in Saccharomyces cerevisiae from vectors utilizing the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Gene. 1984;32:263–274. doi: 10.1016/0378-1119(84)90002-7. [DOI] [PubMed] [Google Scholar]
- 6.Björkqvist S., Ansell R., Adler L., Lidén G. Physiological response to anaerobicity of glycerol-3-phosphate dehydrogenase mutants of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1997;63:128–132. doi: 10.1128/aem.63.1.128-132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlquist M., Fernandes R.L., Helmark S., Heins A.L., Lundin L., Sørensen S.J., Gernaey K.V., Lantz A.E. Physiological heterogeneities in microbial populations and implications for physical stress tolerance. Microb. Cell Fact. 2012;11:94. doi: 10.1186/1475-2859-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlquist M., Gibson B., Karagul Yuceer Y., Paraskevopoulou A., Sandell M., Angelov A.I., Gotcheva V., Angelov A.D., Etschmann M., de Billerbeck G.M., Liden G. Process engineering for bioflavour production with metabolically active yeasts - a mini-review. Yeast. 2014;32:123–143. doi: 10.1002/yea.3058. [DOI] [PubMed] [Google Scholar]
- 9.Cormack B.P., Bertram G., Egerton M., Gow N.A., Falkow S., Brown A.J. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology. 1997;143(Pt 2):303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 10.Ehsani M., Fernandez M.R., Biosca J.A., Julien A., Dequin S. Engineering of 2,3-butanediol dehydrogenase to reduce acetoin formation by glycerol-overproducing, low-alcohol Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2009;75:3196–3205. doi: 10.1128/AEM.02157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eriksson P., Andre L., Ansell R., Blomberg A., Adler L. Cloning and characterization of GPD2, a second gene encoding sn-glycerol 3-phosphate dehydrogenase (NAD+) in Saccharomyces cerevisiae, and its comparison with GPD1. Mol. Microbiol. 1995;17:95–107. doi: 10.1111/j.1365-2958.1995.mmi_17010095.x. [DOI] [PubMed] [Google Scholar]
- 12.Etcheverry T. Induced expression using yeast copper metallothionein promoter. Methods Enzymol. 1990;185:319–329. doi: 10.1016/0076-6879(90)85028-m. [DOI] [PubMed] [Google Scholar]
- 13.Fernandes R.L., Carlquist M., Lundin L., Heins A.L., Dutta A., Sorensen S.J., Jensen A.D., Nopens I., Lantz A.E., Gernaey K.V. Cell mass and cell cycle dynamics of an asynchronous budding yeast population: experimental observations, flow cytometry data analysis, and multi-scale modeling. Biotechnol. Bioeng. 2013;110:812–826. doi: 10.1002/bit.24749. [DOI] [PubMed] [Google Scholar]
- 14.Fernandes R.L., Nierychlo M., Lundin L., Pedersen A.E., Puentes Tellez P.E., Dutta A., Carlquist M., Bolic A., Schapper D., Brunetti A.C., Helmark S., Heins A.L., Jensen A.D., Nopens I., Rottwitt K., Szita N., van Elsas J.D., Nielsen P.H., Martinussen J., Sorensen S.J., Lantz A.E., Gernaey K.V. Experimental methods and modeling techniques for description of cell population heterogeneity. Biotechnol. Adv. 2011;29:575–599. doi: 10.1016/j.biotechadv.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Ferrer-Miralles N., Domingo-Espin J., Corchero J.L., Vazquez E., Villaverde A. Microbial factories for recombinant pharmaceuticals. Microb. Cell Fact. 2009;8 doi: 10.1186/1475-2859-8-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Förster J., Famili I., Fu P., Palsson B.O., Nielsen J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerngross T.U. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotechnol. 2004;22:1409–1414. doi: 10.1038/nbt1028. [DOI] [PubMed] [Google Scholar]
- 18.Gietz R.D., Woods R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 19.Guadalupe-Medina V., Metz B., Oud B., van Der Graaf C.M., Mans R., Pronk J.T., van Maris A.J. Evolutionary engineering of a glycerol-3-phosphate dehydrogenase-negative, acetate-reducing Saccharomyces cerevisiae strain enables anaerobic growth at high glucose concentrations. Microb. Biotechnol. 2014;7:44–53. doi: 10.1111/1751-7915.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn-Hägerdal B., Karhumaa K., Jeppsson M., Gorwa-Grauslund M.F. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 2007;108:147–177. doi: 10.1007/10_2007_062. [DOI] [PubMed] [Google Scholar]
- 21.Hebisch E., Knebel J., Landsberg J., Frey E., Leisner M. High variation of fluorescence protein maturation times in closely related E. coli strains. PloS One. 2013;8 doi: 10.1371/journal.pone.0075991. e75991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue H., Nojima H., Okayama H. High efficiency transformation of E. coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 23.Inouye S., Tsuji F.I. Evidence for redox forms of the Aequorea green fluorescent protein. FEBS Lett. 1994;351:211–214. doi: 10.1016/0014-5793(94)00859-0. [DOI] [PubMed] [Google Scholar]
- 24.Ishii J., Kondo T., Makino H., Ogura A., Matsuda F., Kondo A. Three gene expression vector sets for concurrently expressing multiple genes in Saccharomyces cerevisiae. FEMS Yeast Res. 2014;14:399–411. doi: 10.1111/1567-1364.12138. [DOI] [PubMed] [Google Scholar]
- 25.Jeffries T.W., Jin Y.S. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl. Microbiol. Biotechnol. 2004;63:495–509. doi: 10.1007/s00253-003-1450-0. [DOI] [PubMed] [Google Scholar]
- 26.Knudsen J.D., Carlquist M., Gorwa-Grauslund M. NADH-dependent biosensor in Saccharomyces cerevisiae: principle and validation at the single cell level. AMB Expr. 2014;4(1) doi: 10.1186/s13568-014-0081-4. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lidén G., Walfridsson M., Ansell R., Anderlund M., Adler L., Hahn-Hägerdal B. A glycerol-3-phosphate dehydrogenase-deficient mutant of Saccharomyces cerevisiae expressing the heterologous XYL1 gene. Appl. Environ. Microbiol. 1996;62:3894–3896. doi: 10.1128/aem.62.10.3894-3896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mumberg D., Muller R., Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mumberg D., Müller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 30.Musti A.M., Zehner Z., Bostian K.A., Paterson B.M., Kramer R.A. Transcriptional mapping of two yeast genes coding for glyceraldehyde 3-phosphate dehydrogenase isolated by sequence homology with the chicken gene. Gene. 1983;25:133–143. doi: 10.1016/0378-1119(83)90175-0. [DOI] [PubMed] [Google Scholar]
- 31.Nissen T.L., Hamann C.W., Kielland-Brandt M.C., Nielsen J., Villadsen J. Anaerobic and aerobic batch cultivations of Saccharomyces cerevisiae mutants impaired in glycerol synthesis. Yeast. 2000;16:463–474. doi: 10.1002/(SICI)1097-0061(20000330)16:5<463::AID-YEA535>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 32.Nissen T.L., Schulze U., Nielsen J., Villadsen J. Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology. 1997;143(Pt 1):203–218. doi: 10.1099/00221287-143-1-203. [DOI] [PubMed] [Google Scholar]
- 33.Norbeck J., Pahlman A.K., Akhtar N., Blomberg A., Adler L. Purification and characterization of two isoenzymes of dl-glycerol-3-phosphatase from Saccharomyces cerevisiae. Identification of the corresponding GPP1 and GPP2 genes and evidence for osmotic regulation of Gpp2p expression by the osmosensing mitogen-activated protein kinase signal transduction pathway. J. Biol. Chem. 1996;271:13875–13881. doi: 10.1074/jbc.271.23.13875. [DOI] [PubMed] [Google Scholar]
- 35.Overkamp K.M., Bakker B.M., Kotter P., van Tuijl A., de Vries S., van Dijken J.P., Pronk J.T. In vivo analysis of the mechanisms for oxidation of cytosolic NADH by Saccharomyces cerevisiae mitochondria. J. Bacteriol. 2000;182:2823–2830. doi: 10.1128/jb.182.10.2823-2830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Partow S., Siewers V., Bjorn S., Nielsen J., Maury J. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010;27:955–964. doi: 10.1002/yea.1806. [DOI] [PubMed] [Google Scholar]
- 37.Påhlman I.L., Gustafsson L., Rigoulet M., Larsson C. Cytosolic redox metabolism in aerobic chemostat cultures of Saccharomyces cerevisiae. Yeast. 2001;18:611–620. doi: 10.1002/yea.709. [DOI] [PubMed] [Google Scholar]
- 38.Paalme T., Kahru A., Elken R., Vanatalu K., Tiisma K., Raivo V. The computer-controlled continuous culture of Escherichia coli with smooth change of dilution rate (A-stat) J. Microbiol. Methods. 1995;24:145–153. [Google Scholar]
- 39.Redden H., Morse N., Alper H.S. The synthetic biology toolbox for tuning gene expression in yeast. FEMS Yeast Res. 2015;15:1–10. doi: 10.1111/1567-1364.12188. [DOI] [PubMed] [Google Scholar]
- 40.Regenberg B., Grotkjaer T., Winther O., Fausboll A., Akesson M., Bro C., Hansen L.K., Brunak S., Nielsen J. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 2006;7:R107. doi: 10.1186/gb-2006-7-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roldão A., Kim I.-K., Nielsen J. Bridging omics technologies with synthetic biology in yeast industrial biotechnology. In: Wittmann C., Lee S.Y., editors. Systems Metabolic Engineering. Springer; Netherlands: 2012. pp. 271–327. [Google Scholar]
- 42.Roostalu J., Joers A., Luidalepp H., Kaldalu N., Tenson T. Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol. 2008;8:68. doi: 10.1186/1471-2180-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrock J., Russell D.W. Cold Spring Harbor Laboratory Press; New York, USA: 2001. Molecular Cloning, A Laboratory Manual. [Google Scholar]
- 44.Sandström A.G., Almqvist H., Portugal-Nunes D., Neves D., Lidén G., Gorwa-Grauslund M.F. Saccharomyces cerevisiae: a potential host for carboxylic acid production from lignocellulosic feedstock? Appl. Microbiol. Biotechnol. 2014;98:7299–7318. doi: 10.1007/s00253-014-5866-5. [DOI] [PubMed] [Google Scholar]
- 45.Skorupa Parachin N., Carlquist M., Gorwa-Grauslund M.F. Comparison of engineered Saccharomyces cerevisiae and engineered Escherichia coli for the production of an optically pure keto alcohol. Appl. Microbiol. Biotechnol. 2009;84:487–497. doi: 10.1007/s00253-009-1964-1. [DOI] [PubMed] [Google Scholar]
- 46.Stewart J.D. Organic transformations catalyzed by engineered yeast cells and related systems. Curr. Opin. Biotechnol. 2000;11:363–368. doi: 10.1016/s0958-1669(00)00111-7. [DOI] [PubMed] [Google Scholar]
- 47.Sun J., Shao Z., Zhao H., Nair N., Wen F., Xu J.H. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2012;109:2082–2092. doi: 10.1002/bit.24481. [DOI] [PubMed] [Google Scholar]
- 48.Valadi H., Larsson C., Gustafsson L. Improved ethanol production by glycerol-3-phosphate dehydrogenase mutants of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1998;50:434–439. doi: 10.1007/s002530051317. [DOI] [PubMed] [Google Scholar]
- 49.Valkonen M., Mojzita D., Penttilä M., Bencina M. Noninvasive high-throughput single-cell analysis of the intracellular pH of Saccharomyces cerevisiae by ratiometric flow cytometry. Appl. Environ. Microbiol. 2013;79:7179–7187. doi: 10.1128/AEM.02515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Dijken J.P., Bauer J., Brambilla L., Duboc P., Francois J.M., Gancedo C., Giuseppin M.L., Heijnen J.J., Hoare M., Lange H.C., Madden E.A., Niederberger P., Nielsen J., Parrou J.L., Petit T., Porro D., Reuss M., van Riel N., Rizzi M., Steensma H.Y., Verrips C.T., Vindelov J., Pronk J.T. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 2000;26:706–714. doi: 10.1016/s0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
- 51.van Dijken J.P., Scheffers W.A. Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol. Lett. 1986;32:199–224. [Google Scholar]
- 52.Verduyn C., Postma E., Scheffers W.A., Van Dijken J.P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 53.Walsh G. Biopharmaceutical benchmarks 2010. Nat. Biotechnol. 2010;28:917–924. doi: 10.1038/nbt0910-917. [DOI] [PubMed] [Google Scholar]
- 54.Weber N., Gorwa-Grauslund M., Carlquist M. Exploiting cell metabolism for biocatalytic whole-cell transamination by recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2014;98:4615–4624. doi: 10.1007/s00253-014-5576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weusthuis R.A., Lamot I., van der Oost J., Sanders J.P. Microbial production of bulk chemicals: development of anaerobic processes. Trends Biotechnol. 2011;29:153–158. doi: 10.1016/j.tibtech.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 56.Weusthuis R.A., Visser W., Pronk J.T., Scheffers W.A., van Dijken J.P. Effects of oxygen limitation on sugar metabolism in yeasts: a continuous-culture study of the Kluyver effect. Microbiology. 1994;140(Pt 4):703–715. doi: 10.1099/00221287-140-4-703. [DOI] [PubMed] [Google Scholar]
- 57.Knudsen J.D. Lund University; 2015. An NADH-Coupled Biosensor for Engineering Redox Metabolism in Saccharomyces cerevisiae, Doctoral Thesis. [Google Scholar]