Highlights
-
•
A scFv intrabody specific for the NPMc+ mutant NES sequence was isolated.
-
•
It was expressed as a fusion with a NLS and such construct accumulates in the nucleus.
-
•
The scFv-NLS fusion binds its antigen in the cytoplasm of eukaryotic cells.
-
•
The complex shuttles to the nucleus but accumulates in the cytoplasm.
-
•
Stronger NLS should be developed to revert the strength of pathogenic NES.
Abbreviations: AML, acute myeloid leukemia; CRM1, Chromosomal Region Maintenance 1; GST, glutathione S-transferase; MBP, maltose binding protein; NES, nuclear export signal; NLS, nuclear localization signal; NPM1, nucleophosmin; NPMc+, cytoplasmic nucleophosmin; scFv, single-chain variable fragment
Keywords: Intrabody, Leukemia, Protein shuttling, Recombinant fusion protein, Single-chain antibody
Abstract
The cytoplasmic accumulation of NPM1 (NPMc+) is found in acute myeloid leukemia (AML) with NPM1 mutation. NPM1 must shuttle between nucleus and cytoplasm to assure physiological protein synthesis and, therefore, the elimination of NPMc+ is not a suitable therapeutic option. We isolated, characterized, and produced a functional scFv intrabody fused to nuclear localization signal(s) (NLS) that does not recognize NPM1 but binds to the mutant-specific C-terminal NES (nuclear export signal) of NPMc+, responsible for its cytoplasmic accumulation. The scFv-NLS fusion accumulated in the nuclei of wild type cells and strongly bound to its antigen in the cytoplasm of NPMc+ expressing cells. However, it failed to relocate the majority of NPMc+ in the nucleus, even when fused to four NLS. Our results show the technical feasibility of producing recombinant intrabodies with defined sub-cellular targeting and nuclear accumulation but the lack of information concerning the features that confer variable strength to the signal peptides impairs the development of biomolecules able to counteract pathological sub-cellular distribution of shuttling proteins.
1. Introduction
Nucleophosmin (NPM1) is a nucleolar multifunctional phosphoprotein involved in RNA metabolism [1], [2], [3], regulation of the p19/ARF-p53 tumor-suppressor pathway [4], [5] and c-Myc turnover through Fbw7γ [6]. Under physiological conditions, the protein shuttles between nucleus and cytoplasm. In about one-third of adult patients with AML with normal karyotype, it has been demonstrated that AML cells bear mutations in the last coding exon of the NPM1 gene (exon 12) [7], [8], [9]. More than 40 heterozygous different mutations have been described. The mutations result in frame shift and the loss of the two tryptophan residues located in the C-terminal portion of the protein that are necessary for nucleolar localization. The insertion of short nucleotide stretches of eleven amino acids generates the de novo formation of a Chromosomal Region Maintenance 1 (CRM1)/Exportin 1-dependent NES responsible for mutant NPM1 cytoplasmic delocalization (NPMc+) [10], [11], [12]. Although a correlation between NPM1 cytoplasmic accumulation and leukemia initiation and progression has been recently demonstrated in vivo in murine models [13], [14], so far there is no direct molecular evidence of the mechanism by which NPMc+ can induce pathological conditions. It has been suggested that NPMc+ could form hetero-octamers with NPM1 inducing its delocalization and that of proteins normally associated to NPM1, such as p19/ARF and Fbw7γ [4], [5], [6], [15]. A monoclonal antibody (T26) specific for the cytoplasmic mutation has been demonstrated helpful to confirm the connection between NPMc+ expression and AML in patients [16]. However, when we performed a double staining to identify both NPM1 and NPMc+ localization, it turned out that a significant portion of the wild type protein was still located in the nucleoli [17], questioning the hypothesis of a massive NPM1 migration to the cytoplasm. Nevertheless, both the shuttling and the residential activities of NPM1 are necessary for the normal metabolism since NPM1 seems to be the rate-limiting nuclear export shuttle for ribosome components in mammalian cells and an indispensable regulator of protein synthesis [18]. The diminished NPM1 shuttling capacity impairs the regular ribosome assembly, places genetic pressure upon p19/ARF/p53 pathway, and leads to mutations resulting in cellular transformation [18]. This means that NPM1 shuttling must be preserved as well as its predominant nucleolar accumulation. Consequently, any therapeutic perspective should re-establish this equilibrium rather than inactivate NPMc+ by means of neutralizing drugs.
Intrabodies have been successfully used in the past to knock-out their targets or sequester their antigen in specific sub-cellular compartments [19], [20], [21]. Similarly, we isolated a scFv antibody specific for the de novo exclusive NES motif present in the mutated NPMc+, confirmed its correct folding when it was expressed as an intrabody, and fused it to a sequence corresponding to a repeat of nuclear localization signals (NLS). Despite the effective binding to NPMc+ and the transient relocation into the nucleus, our data showed that the antigen–antibody complex remained statistically localized in the cytoplasm, a result that seems to confirm some previous reports underlining the large efficiency variability existing among nuclear localization signal peptides [22], [23].
2. Materials and methods
2.1. Expression and purification of recombinant NPMc+
Full-length NPMc+ was expressed as a GST (glutathione S-transferase) fusion from pGEX4T vector and purified by affinity chromatography [24] using GSTrapFF column and ÄKTA Explorer (GE Healthcare). The C-terminal NPMc+ fragment corresponding to the 45 amino acids from 255 to 298 was synthesized by PCR, cloned in pETM44 vector [25] as MBP (maltose binding protein)-6× His tag fusion and transformed in BL21 cells. Cultures were grown in ZYP-5052 auto-inducing medium [26]. Purification was performed combining HisTrapHP column and ÄKTA Explorer (GE Healthcare).
2.2. Selection and subcloning of phage displayed scFvs
Human monoclonal scFv antibodies specific to NPMc+ were isolated from the synthetic ETH-2 Gold phage display library [27]. A pre-panning incubation step of the library against MBP at a concentration of 100 μg mL−1 was performed before each panning round to deplete anti-MBP binders. Three rounds of panning were performed on Nunc-Immuno™ Maxisorp™ tubes (Nunc) coated with the fusion construct NPMc+–MBP at a concentration of 25 μg mL−1 in 50 mM sodium carbonate buffer, pH 9.6 [28] and scFvs were screened by ELISA [27]. Six clones with an absorbance value higher than 0.49 and negative for the fusion tag were considered positive (Supplementary Fig. 1A) and sequenced using the following primers: Fdseq1 5′-GAATTTTCTGTATGAGG-3′ and PelbBack 5′-AGCCGCTGGATTGTTATTAC-3′. The results indicated that all the six clones shared the same sequence, suggesting a high selective pressure toward one specific binder (Supplementary Fig. 1B). It was produced in large scale in TG1 cells and purified on HiTrapMabSelectSuRE ProteinA column followed by size exclusion chromatography on HiLoad 16/60 Superdex 200 using ÄKTA Explorer (GE Healthcare). The mouse anti-Myc monoclonal antibody 9E10 (8 μg mL−1) was used as a primary antibody in ELISA test. The NLS corresponding to the SV40 large T-antigen was fused to scFv by PCR using the following primers: FW: 5′-CCAAGCTTCCATGGAGGTGCAGCTGTTGGAGTCTGGG-3′; REV: 5′CTAGGCGC GGCCGCATACCCCT ACGACGTGCCCGACTACCCCAAAAAGAAACGAAAAGTA TAGTCTAGACTAG-3′ and the product was cloned HindIII-XbaI in the pcDNA3.1 vector (Invitrogen) to obtain NLS-HA fusions. The same approach was used to insert till to four SV40 NLS sequences. Flag tag and GFP-scFv fusion were obtained by cloning HindIII-XbaI the antibody cDNA in the pcDNA3.1 and in the pEGFP-C1 (Clontech) vectors, respectively.
Supplementary Fig. S1 related to this article can be found, in the online version, at doi:10.1016/j.btre.2014.05.008.
scFv specificity for NPMc+. (A) ELISA test. Absorbance values were measured at 405 nm using scFv-expressing bacterial supernatants in combination with either NPMc+-MBP fusion fragment or MBP alone. (B) Sequence of the VH and VL domains of the selected anti-NPMc+ scFv antibody. (C) The protein fractions corresponding to the purified constructs of the NPMc+ fragment 255–298 fused to either MBP (NPMc+ fragment-MBP, lane 2) or the full-length NPMc+ mutant fused to GST (NPMc+-GST, lane 6), were separated by SDS-PAGE gel in parallel with purified MBP (lane 1), GST (lane 5), and the total lysate recovered from either non-transfected insect cells (lane 3) or insect cells expressing GFP (lane 4), the mutant NPMc+ (lane 7), and the wild type NPM1 (lane 8). Protein bands were identified by western immuno-blot using scFv-containing cell culture supernatant in combination with mouse anti-Myc monoclonal antibody (9E10). (D) Insect cell lysates expressing either NPMc+ (lane 1) or wild type NPM1 (lane 2) were separated by SDS-PAGE gel and probed with mouse monoclonal antibodies specific for either the wild type NPM1 C-terminal end (338) or for the common N-terminal region of NPM (376).
2.3. Cell culture and transfection
HeLa cells were grown in Dulbecco's modified Eagle Medium (Lonza) supplemented with 10% FBS, l-glutamine (2 mM), penicillin (100 U mL−1), streptomycin (100 mg mL−1). OCI-AML2 and OCI-AML3 [29] cell lines were grown in MEM Alpha + GlutaMAX™-I medium (Gibco) supplemented with 20% FBS, glutamine and antibiotics. Transient transfections were performed using Lipofectamine™ 2000 (Invitrogen). Sf9 (Spodoptera frugiperda) insect cells were cultured at 27 °C in Sf 900 II SMF medium (Gibco) and transfected with pFastBacDual plasmids (Invitrogen) expressing either wild type NPM1 or NPMc+ using Insectogene T030-1.0 (Biontex). Baculoviral supernatant was collected after 96 h and used for two cycles of infection.
2.4. Immunoblotting and immunoprecipitation
For immunoprecipitation, cells were lysed in 50 mM Tris–HCl, pH 8, 150 mM NaCl, 0.5% NP40, and protease inhibitors. Ten micrograms of scFv were added overnight at 4 °C to HeLa and OCI-AML3 cell lysates followed by protein A/G-sepharose (GE Healthcare). For co-immunoprecipitation experiments, total cell lysate was incubated with mouse M2 anti-Flag agarose beads (Sigma) and with anti-mouse IgG agarose beads (Sigma) for 4 h at 4 °C. Precipitated recombinant purified proteins and cell lysates were separated by SDS-PAGE gel and immunoblotted over a nitrocellulose membrane (Whatman). After incubation with primary antibodies in 5% skimmed milk, the membrane was incubated with horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibodies (Bio-Rad). Primary antibodies used in WB were mouse monoclonal anti-Myc 9E10, mouse monoclonal anti-Flag M2, mouse monoclonal anti-NPMc+ T26, mouse 338 [30] anti-NPM1 C-terminal end, mouse 376 [30] anti-NPM N-terminal region, and supernatant from recombinant scFv. ECL Plus was used as a substrate for chemiluminescent-based protein immune detection (Pierce). Primary antibodies used in IP were mouse monoclonal anti-NPMc+ T26 and recombinant scFv.
2.5. Immunofluorescence assay
Cells grown on cover slips were fixed in paraformaldehyde, washed twice in PBS, permeabilized 5 min in 0.2% Triton X-100, washed again in PBS and blocked in 2% BSA for 30 min at room temperature. Slides were incubated 1 h in blocking buffer containing primary antibodies, washed extensively in PBS, and incubated with CY3-conjugated donkey anti-mouse immunoglobulin (Jackson ImmunoResearch) for 30 min. After washing, slides were counterstained with DAPI, rinsed in distilled water, mounted with mowiol, and assessed at the DAPI, GFP and CY3 channels. Images were acquired using an Olympus AX70 microscope equipped with a CoolSNAP EZ Turbo 1394 camera (Photometrics) and processed using ImageJ 1.43 software (Wayne Rasband, NIH). Leptomycin B experiments were performed as previously described [10]. Confocal microscopy was performed on a Leica TCS SP5 equipped with violet (405 nm) and blue (488 nm) excitation laser lines. Primary antibodies used for IF were mouse monoclonal anti-Myc 9E10, mouse monoclonal anti-HA, mouse monoclonal anti-NPMc+ T26, and recombinant scFv.
3. Results
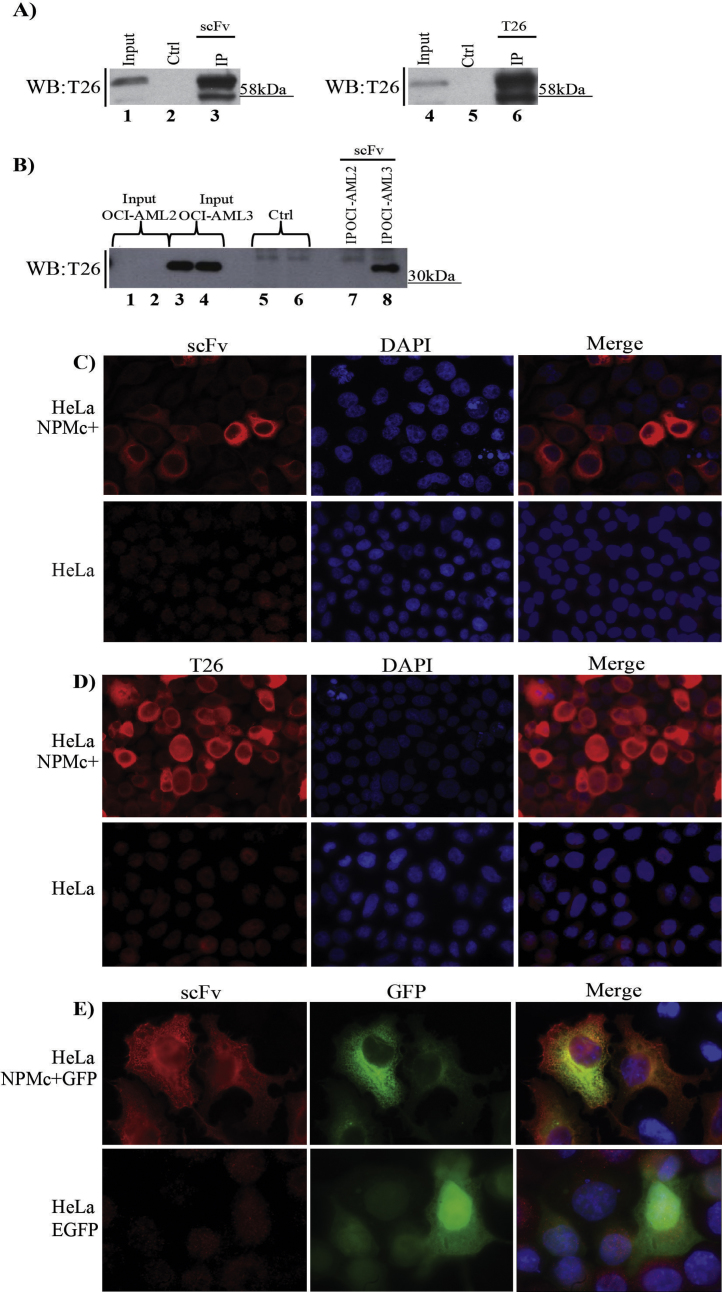
Panning the synthetic ETH-2 Gold phage display library [27] against the C-terminal peptide of the NPMc+ mutant succeeded in isolating some scFvs that specifically bound to the nuclear export signal (NES) sequence responsible for the strong cytoplasmic localization of the target protein (Supplementary Fig. 1). The antibody fragment identified among the positive clones (Supplementary Fig. 1B) was produced as a stable molecule and was chosen for further characterization. As shown by western immuno-blot analysis (Supplementary Fig. 1C), the antibody recognized its recombinant antigen alone as well as fused to either MBP or GST, while no signal was detected in the presence of the carrier proteins and of the control recombinant proteins GFP and NPM1. Similarly, the scFv detected the NPMc+ isoform expressed in insect cells with the same specificity of monoclonal antibodies (Supplementary Fig. 1D) and successfully pulled-down NPMc+ from total cell lysates of both NPMc+-transfected HeLa cells (Fig. 1A) and human acute myeloid leukemia OCI-AML3 cells that constitutively express NPMc+ (Fig. 1B). As expected, it did not immuno-precipitate NPM1 from human acute myeloid leukemia OCI-AML2 cells in which NPMc+ is not expressed. Pull-down efficiency was comparable to that of the anti-NPMc+ T26 monoclonal antibody [16]. The two antibodies visualized the same NPMc+ pattern distribution in HeLa cells, although the monovalent scFv apparently bound the target protein with lower avidity than the bivalent monoclonal antibody (Fig. 1C and D). Finally, the scFv co-localized with the cytoplasmically expressed NPMc+ in transiently transfected cells, as shown by immunofluorescence assay (Fig. 1E).
Fig. 1.
Immunospecificity of the anti-NPMc+ scFv. (A) HeLa cell lysates transiently expressing NPMc+-GFP were used for immunoprecipitation with the anti-NPMc+ antibodies: scFv (lanes 1–3) and T26 (lanes 4–6). Control experiments (lanes 2 and 5) were performed with protein A-sepharose and protein G-sepharose, respectively. Membranes were probed with T26 mouse monoclonal antibody. (B) scFv was used to immunoprecipitate NPMc+ from OCI-AML2 and OCI-AML3 cell lysates (lanes 7 and 8). Lanes 1 and 2: inputs of OCI-AML2 cell lysate; lanes 3 and 4: inputs of OCI-AML 3 cell lysate. As controls, protein A-sepharose was incubated alone in the presence of OCI-AML2 (lane 5) and OCI-AML3 (lane 6) cell lysates. The membrane was probed with T26 mouse monoclonal antibody. Immunofluorescence assay on HeLa cells transiently expressing NPMc+-FlagHA (upper panels) and on not transfected HeLa cells (lower panels) was performed using purified scFv (C) and T26 mouse monoclonal antibody (D). (E) Immunofluorescence assay on HeLa cells transiently expressing either NPMc+-GFP (upper panels) or EGFP (lower panels) was performed using the purified scFv antibody. Fusion to GFP did not impair physiological NPMc+ localization [15].
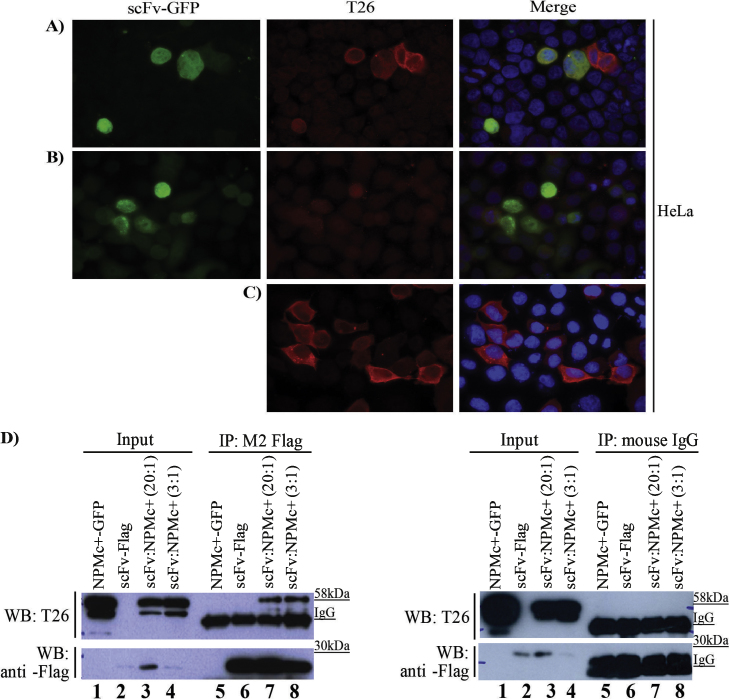
These preliminary data confirmed that the scFv was a reliable binder of the NPMc+ mutant and therefore we evaluated the possibility to express it as an intrabody in HeLa cell cytoplasm. HeLa cells were transiently co-transfected with NPMc+ and a scFv-GFP fusion. The frequency of cells co-expressing both constructs was always low (about 5%) but the homogeneous accumulation of green fluorescent (scFv-fusion) protein seems to indicate that the anti-NPMc+ antibody did not aggregate and that it mainly co-localized with its antigen in the cytoplasm (Fig. 2A–C). Similar results were obtained by infecting leukemic cells with retroviral and lentiviral vectors expressing the scFv (data not shown). The immunoprecipitation results (Fig. 2D) confirmed that, upon transient co-expression, the scFv-Flag construct was functionally folded and effectively interacted with its antigen in the intracellular milieu, although at a low stoichiometic ratio.
Fig. 2.
Functional characterization of the isolated anti-NPMc+ scFv. Immunofluorescence assay on HeLa cells transiently co-expressing both scFv-GFP and NPMc+ (A), or scFv-GFP (B) and NPMc+ (C) alone. All the experiments were performed using T26 mouse monoclonal antibody to localize NPMc+. (D) HeLa cells were first transiently co-transfected with the scFv-Flag and the NPMc+-GFP expression vectors. Successively, the Flag-tag was used for affinity precipitation and the resulting material was analyzed by western immuno-blot using T26 mouse monoclonal antibody and mouse anti-Flag M2 antibody, as indicated (left panel). Immunoprecipitation with mouse IgG agarose beads was used as a negative control (right panel). Lanes 1 and 2: inputs of HeLa cells transiently transfected with NPMc+-GFP (lane 1) or scFv-Flag (lane 2). Lanes 3 and 4: inputs of HeLa cells transiently co-transfected with scFv-Flag and NPMc+-GFP at different stoichiometric plasmid ratios (lane 3, scFv-Flag to NPMc+-GFP plasmid ratio of 20:1; lane 4, scFv-Flag to NPMc+-GFP plasmid ratio of 3:1). Lanes 5–8: corresponding immunoprecipitations.
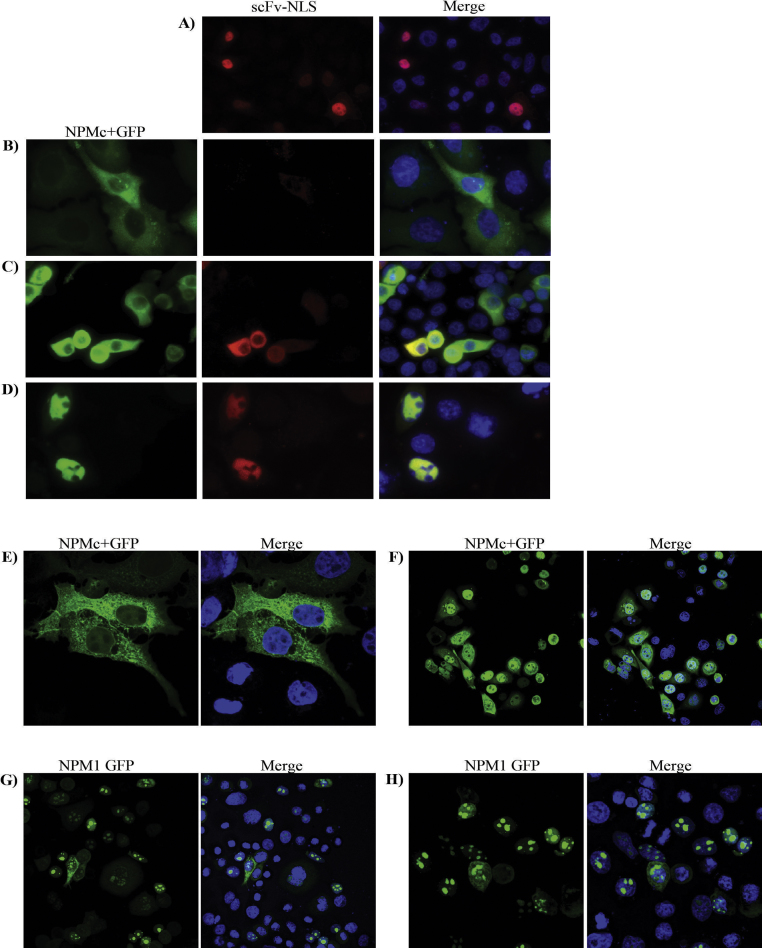
Summarizing, the scFv specific for the C-terminus of the mutated NPMc+ could be expressed in the cytoplasm of mammalian cells as a functional intrabody. Consequently, we prepared a reagent composed by the fusion of the recombinant antibody together with a NLS to evaluate the possibility to bind the cytoplasmic NPMc+ and relocate it into the nucleus. The scFv-NLS construct effectively accumulated into the nucleus (Fig. 3A) and co-accumulated with NPMc+ in the same compartment when the protein nuclear export was inhibited by treating the cells with leptomycin B, a CRM1-dependent nuclear export inhibitor (Fig. 3D). In the absence of leptomycin B treatment, the scFv failed to relocate the cytoplasmic mutant NPMc+ (Fig. 3B) and we observed rather the opposite, namely the antigen sequestered the antibody in the cytoplasm (Fig. 3C). The fusion of four NLS to the scFv did not modify the equilibrium (data not shown). Confocal microscopy imaging showed that NPMc+-GFP (Fig. 3E) accumulated very rapidly in the nuclei of leptomycin B-treated cells even in the absence of scFv-NLS (Fig. 3F). The leptomycin B-dependent nuclear accumulation of NPMc+ and NPM1 in the nucleus was equally effective after 1 h (Fig. 3G and H) although the NPM1 protein accumulation was faster (data not shown). The relatively rapid accumulation of NPMc+ in the nucleus and the rare availability of co-transfected cells impaired to demonstrate a statistically significant contribution of scFv-NLS to the protein nuclear uptake (data not shown).
Fig. 3.
Sub-cellular localization of the scFv-NLS – NPMc+-GFP complex in transiently co-transfected HeLa cells. ScFv-NLS (A) and NPMc+-GFP (B) transfected alone were used as controls. scFv-NLS and NPMc+-GFP co-localize in transiently co-transfected HeLa cells (C). The expression of scFv-NLS was detected by anti-HA mouse monoclonal antibody. NPMc+-GFP and scFv-NLS co-accumulate in the nucleus (D) when cells are treated with leptomycin B, an inhibitor of protein nuclear export. Nevertheless, confocal microscopy imaging allows observing that NPMc + -GFP (E) accumulates in the nuclei of leptomycin B-treated cells very rapidly, even in the absence of scFv-NLS (F). Wild-type NPM1 nucleolar localization before (G) and after leptomycin B treatment (H).
4. Discussion
Sub-cellular localization of proteins shuttling between nucleus and cytoplasm is the consequence of the dynamic equilibrium determined by the relative strength of the two opposite fluxes. In the case of NPM1, both NLS and NES putative motifs are embedded into the wild type sequence, as expected for a protein physiologically shuttling between nucleus and cytoplasm. However, it is not yet clear how many different signal motifs contribute to tune the shuttling, due to the fact that both the leucine residues located at positions 42 and 44 as well as the region between amino acids 92–104 seem to be essential for the export [31], [32]. On the other hand, the two tryptophan residues located at amino acids 288 and 290 display a synergic nucleolar localization effect [11] additive to the contribution of the canonical NLS sequences positioned at 152–157 and 190–197 [33]. Different mutations generate a de novo C-terminal NES leading to a complete shift of the equilibrium toward the cytoplasmic accumulation. At the same time, the inhibition tests in the presence of leptomycin B confirm that the situation remains highly dynamic, since the NPMc+ is relocated to the nucleus in less than 1 h. These observations suggest that NPMc+ is not sequestered in the cytoplasm, that the apparent lack of relocalization is due to unequal rates of translocation in the two directions, and that a modification between the relative speed of export and import might re-establish a preferential nuclear accumulation. It has been recently reported that CRM1 overexpression modifies this equilibrium and correlates with metastasis and poor prognosis in different human cancers [34]. However, despite some positive pre-clinical indications [35], [36], this transporter controls the shuttling of too many essential proteins to be considered an ideal therapeutic target. As an alternative possibility, the equilibrium between the two opposite fluxes could be modified by acting on the strength of the import/export motifs, as it happens in some pathological conditions [37].
The therapeutic potential of tuning the protein delivery to suitable sub-cellular compartments by means of intrabodies have been recently reviewed [21], [38]. Conventional IgGs do not fold correctly in the reducing cytoplasmic milieu but recombinant antibody fragments with simpler structures can reach their functional conformation despite the unfavorable redox conditions. In basic research, the effect of the rapid removal of cytoplasmic proteins has been evaluated by using single-domain antibodies that can trap GFP-tagged proteins and deliver them to ubiquitin-dependent degradation [39]. However, protein sub-cellular re-localization is not a straightforward application since artificially introduced sub-cellular localization sequences clash with native and discording signal sequences [40], a condition that can result in unpredictable protein distribution inside the cell [37]. Furthermore, physiological NES and NLS have apparently evolved as “weak” signals, whereas pathological motifs can be extremely more effective. In this perspective, it would be crucial to have models to predict the effect of coexisting signal sequences of different strength and driving to opposite directions. We have shown that it was possible to select an anti-NPMc+ scFv antibody, to express it as a functional intrabody that binds its antigen in human cell cytoplasm, and to obtain its nuclear accumulation after fusion to suitable NLS. Nevertheless, the antibody–antigen complex was not retained in the nucleus probably because of the different efficiencies of the available import and export signal sequences. The mutation-dependent export domain of NPMc+ reverts the predominantly nucleolar localization enabled by the two NLS sequences embedded into the NPM1 sequence. Apparently, even the addition of four NLS sequences to the scFv did not significantly modify the NPMc+ sub-cellular statistical distribution. Insufficient total driving strength and structural hindrance due to the repeats could be responsible for the negative result. Furthermore, the affinity and the dissociation kinetics of the antibody to its antigen could represent two additional crucial factors for the regulation of NPMc+ shuttling. The accessibility of the NPMc+ epitope for the scFv is probably critical for regulating the binding kinetics: too rapid release from its antigen would impair nucleolar import, whereas too strong binding could block NPMc+ export.
Altogether, these data suggest that our strategy of relocating NPMc+ could be feasible whether a suitable NLS, alone or in combination with adaptor proteins [41], would be available to compete with the super-physiological NES. There are very few scientific reports that investigated quantitatively the molecular parameters controlling the effectiveness of leader sequences [22], [42] and no obvious candidate is available for our model. We believe that an effort in discovering leader sequences to tune the delivery of recombinant antibodies with different binding features would be very useful and allow the modulation of protein sub-cellular (re)localization for therapeutic applications.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Author contributions
C.M. performed research and analyzed data; C.S. and D.P. performed research; E.C., P.G.P., and A.dM. designed research and analyzed data, C.M. and A.dM. wrote the manuscript. All the authors have approved the final version of the manuscript.
Acknowledgements
The authors are grateful to S. Bossi and G. Ossolengo for technical support with insect cell culture and protein purifications. This work was supported by Grants from AIRC (Associazione Italiana per la Ricerca sul Cancro) to E.C., P.G.P., and A.d.M.
Footnotes
Available online 27 May 2014
References
- 1.Tarapore P., Shinmura K., Suzuki H., Tokuyama Y. Thr199 phosphorylation targets nucleophosmin to nuclear speckles and represses pre-mRNA processing. FEBS Lett. 2006;580:399–409. doi: 10.1016/j.febslet.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 2.Lindstrom M.S., Zhang Y. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J. Biol. Chem. 2008;283:15568–15576. doi: 10.1074/jbc.M801151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sagawa F., Ibrahim H., Morrison A.L., Wilusz C.J. Nucleophosmin deposition during mRNA 3′ end processing influences poly(A) tail length. EMBO J. 2011;30:3994–4005. doi: 10.1038/emboj.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurki S., Peltonen K., Latonen L., Kiviharju T.M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5:465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- 5.Colombo E., Bonetti P., Lazzerini Denchi E., Martinelli P. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol. Cell. Biol. 2005;25:8874–8886. doi: 10.1128/MCB.25.20.8874-8886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonetti P., Davoli T., Sironi C., Amati B. Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7 gamma. J. Cell Biol. 2008;182:19–26. doi: 10.1083/jcb.200711040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falini B., Mecucci C., Tiacci E., Alcalay M. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 8.Schnittger S., Schoch C., Kern W., Mecucci C. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 9.Falini B., Martelli M.P., Bolli N., Bonasso R. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood. 2006;108:1999–2005. doi: 10.1182/blood-2006-03-007013. [DOI] [PubMed] [Google Scholar]
- 10.Mariano A.R., Colombo E., Luzi L., Martinelli P. Cytoplasmic localization of NPM in myeloid leukemias is dictated by gain-of-function mutations that create a functional nuclear export signal. Oncogene. 2006;25:4376–4380. doi: 10.1038/sj.onc.1209453. [DOI] [PubMed] [Google Scholar]
- 11.Falini B., Bolli N., Shan J., Martelli M.P. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107:4514–4523. doi: 10.1182/blood-2005-11-4745. [DOI] [PubMed] [Google Scholar]
- 12.Falini B., Nicoletti I., Martelli M.F., Mecucci C. Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): biologic and clinical features. Blood. 2007;109:874–885. doi: 10.1182/blood-2006-07-012252. [DOI] [PubMed] [Google Scholar]
- 13.Cheng K., Sportoletti P., Ito K., Clohessy J.G. The cytoplasmic NPM mutant induces myeloproliferation in a transgenic mouse model. Blood. 2010;115:3341–3345. doi: 10.1182/blood-2009-03-208587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vassiliou G.S., Cooper J.L., Rad R., Li J. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat. Genet. 2011;43:470–475. doi: 10.1038/ng.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colombo E., Martinelli P., Zamponi R., Shing D.C. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res. 2006;66:3044–3050. doi: 10.1158/0008-5472.CAN-05-2378. [DOI] [PubMed] [Google Scholar]
- 16.Gruszka A.M., Lavorgna S., Consalvo M.I., Ottone T. A monoclonal antibody against mutated nucleophosmin 1 for the molecular diagnosis of acute myeloid leukemias. Blood. 2010;116:2096–2102. doi: 10.1182/blood-2010-01-266908. [DOI] [PubMed] [Google Scholar]
- 17.Gruszka A.M., Martinelli C., Sparacio E., Pelicci P.G. The concurrent use of N- and C-terminal antibodies anti-nucleophosmin 1 in immunofluorescence experiments allows for precise assessment of its sub-cellular localization in acute myeloid leukaemia patients. Leukemia. 2012;26:159–162. doi: 10.1038/leu.2011.177. [DOI] [PubMed] [Google Scholar]
- 18.Maggi L.B., Kuchenruether M., Dadey D.Y., Schwope R.M. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the mammalian ribosome. Mol. Cell. Biol. 2008;28:7050–7065. doi: 10.1128/MCB.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardinale A., Filesi I., Mattei S., Biocca S. Intracellular targeting and functional analysis of single-chain Fv fragments in mammalian cells. Methods. 2004;34:171–178. doi: 10.1016/j.ymeth.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Beyer F., Doebis C., Busch A., Ritter T. Decline of surface MHC I by adenoviral gene transfer of anti-MHC I intrabodies in human endothelial cells-new perspectives for the generation of universal donor cells for tissue transplantation. J. Gene Med. 2004;6:616–623. doi: 10.1002/jgm.548. [DOI] [PubMed] [Google Scholar]
- 21.Lo A.S., Zhu Q., Marasco W.A. Intracellular antibodies (intrabodies) and their therapeutic potential. Handb. Exp. Pharmacol. 2008;181:343–373. doi: 10.1007/978-3-540-73259-4_15. [DOI] [PubMed] [Google Scholar]
- 22.Engelsma D., Bernad R., Calafat J., Fornerod M. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 2004;23:3643–3652. doi: 10.1038/sj.emboj.7600370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutay U., Güttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 24.de Marco A. Two-step metal affinity purification of double-tagged (NusA-His6) fusion proteins. Nat. Protoc. 2006;1:1538–1543. doi: 10.1038/nprot.2006.289. [DOI] [PubMed] [Google Scholar]
- 25.Dümmler A., Lawrence A.M., de Marco A. Simplified screening for the detection of soluble fusion constructs expressed in E. coli using a modular set of vectors. Microb. Cell Fact. 2005;4:34. doi: 10.1186/1475-2859-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Studier F.W., Rosenberg A.H., Dunn J.J., Dubendorff J.W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 27.Silacci M., Brack S., Schirru G., Mårlind J. Design, construction, and characterization of a large synthetic human antibody phage display library. Proteomics. 2005;5:2340–2350. doi: 10.1002/pmic.200401273. [DOI] [PubMed] [Google Scholar]
- 28.Stoll V.S., Blanchard J.S. Buffers: principles and practice. Methods Enzymol. 1990;182:24–38. doi: 10.1016/0076-6879(90)82006-n. [DOI] [PubMed] [Google Scholar]
- 29.Quentmeier H., Martelli M.P., Dirks W.G., Bolli N. Cell line OCI/AML3 bears exon-12 NPM gene mutation-A and cytoplasmic expression of nucleophosmin. Leukemia. 2005;19:1760–1767. doi: 10.1038/sj.leu.2403899. [DOI] [PubMed] [Google Scholar]
- 30.Cordell J.L., Pulford K.A., Bigerna B., Roncador G. Detection of normal and chimeric nucleophosmin in human cells. Blood. 1999;93:632–642. [PubMed] [Google Scholar]
- 31.Wang W., Budhu A., Forgues M., Wang X.W. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat. Cell Biol. 2005;7:823–830. doi: 10.1038/ncb1282. [DOI] [PubMed] [Google Scholar]
- 32.Yu Y., Maggi L.B., Jr., Brady S.N., Apicelli A.J. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol. 2006;26:3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grisendi F., Mecucci C., Falini B., Pandolfi P.P. Nucleophosmin and cancer. Nat. Rev. Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 34.Turner J.G., Dawson J., Dullivan D.M. Nuclear export of proteins and drug resistance in cancer. Biochem. Pharmacol. 2012;83:1021–1032. doi: 10.1016/j.bcp.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inoue H., Kauffman M., Shacham S., Landesman Y. CRM1 blockade by selective inhibitors of nuclear export attenuates kidney cancer growth. J. Urol. 2012;189:2317–2326. doi: 10.1016/j.juro.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Etchin J., Sanda T., Mansour M.R., Kentsis A. KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. Br. J. Haematol. 2013;161:117–127. doi: 10.1111/bjh.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venuto A., de Marco A. Conflict of interests: multiple signal peptides with diverging goals. J. Cell. Biochem. 2013;114:510–513. doi: 10.1002/jcb.24393. [DOI] [PubMed] [Google Scholar]
- 38.Cardinale C., Biocca S. The potential of intracellular antibodies for therapeutic targeting of protein-misfolding diseases. Trends Mol. Med. 2008;14:373–380. doi: 10.1016/j.molmed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Caussinus E., Kanca O., Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 2012;19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- 40.Piccinin S., Tonin E., Sessa S., Demontis S. A Twist box code of p53 inactivation: Twist box:p53 interaction promotes p53 degradation. Cancer Cell. 2012;22:404–415. doi: 10.1016/j.ccr.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Lott K., Cingolani G. The importin β binding domain as a master regulator of nucleocytoplasmic transport. Biochim. Biophys. Acta. 2011;1813:1578–1592. doi: 10.1016/j.bbamcr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitrousis G., Olia A.S., Walker-Kopp N., Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J. Biol. Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
scFv specificity for NPMc+. (A) ELISA test. Absorbance values were measured at 405 nm using scFv-expressing bacterial supernatants in combination with either NPMc+-MBP fusion fragment or MBP alone. (B) Sequence of the VH and VL domains of the selected anti-NPMc+ scFv antibody. (C) The protein fractions corresponding to the purified constructs of the NPMc+ fragment 255–298 fused to either MBP (NPMc+ fragment-MBP, lane 2) or the full-length NPMc+ mutant fused to GST (NPMc+-GST, lane 6), were separated by SDS-PAGE gel in parallel with purified MBP (lane 1), GST (lane 5), and the total lysate recovered from either non-transfected insect cells (lane 3) or insect cells expressing GFP (lane 4), the mutant NPMc+ (lane 7), and the wild type NPM1 (lane 8). Protein bands were identified by western immuno-blot using scFv-containing cell culture supernatant in combination with mouse anti-Myc monoclonal antibody (9E10). (D) Insect cell lysates expressing either NPMc+ (lane 1) or wild type NPM1 (lane 2) were separated by SDS-PAGE gel and probed with mouse monoclonal antibodies specific for either the wild type NPM1 C-terminal end (338) or for the common N-terminal region of NPM (376).