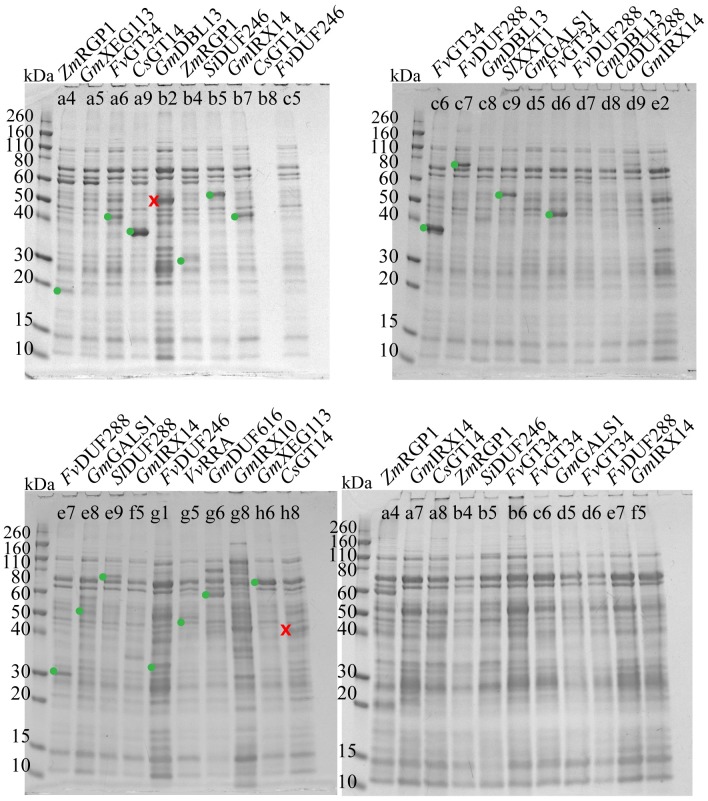
Fig 3. SDS-PAGE of selected samples from the non-Arabidopsis library.
Coomassie-stained gels showing nickel affinity eluates of samples selected from the automated capillary electrophoresis procedure as described. Each lane is marked with the organism and protein name, as well as with the well number for reference to S6 Data. Bands that were excised for mass spectrometry are marked with a green dot (confirmed by mass spectrometry) or a red cross (not confirmed). The lower right gel has samples expressed in the periplasm (pET22DEST vector, no excisions), while the other three gels have samples expressed in the cytoplasm (pET55DEST vector). Sample b8 (upper left gel) has no detectable background on SDS-PAGE, but produces a normal band pattern in the automated capillary electrophoresis, indicating that the absence of bands on SDS-PAGE is likely due to faulty well loading. Ca = Cicer arietinum, Cs = Cucumis sativa, Fv = Fragaria vesca, Gm = Glycine max, Si = Setaria italica, Sl = Solanum lycopersicum, Vv = Vitis vinifera, Zm = Zea mays.