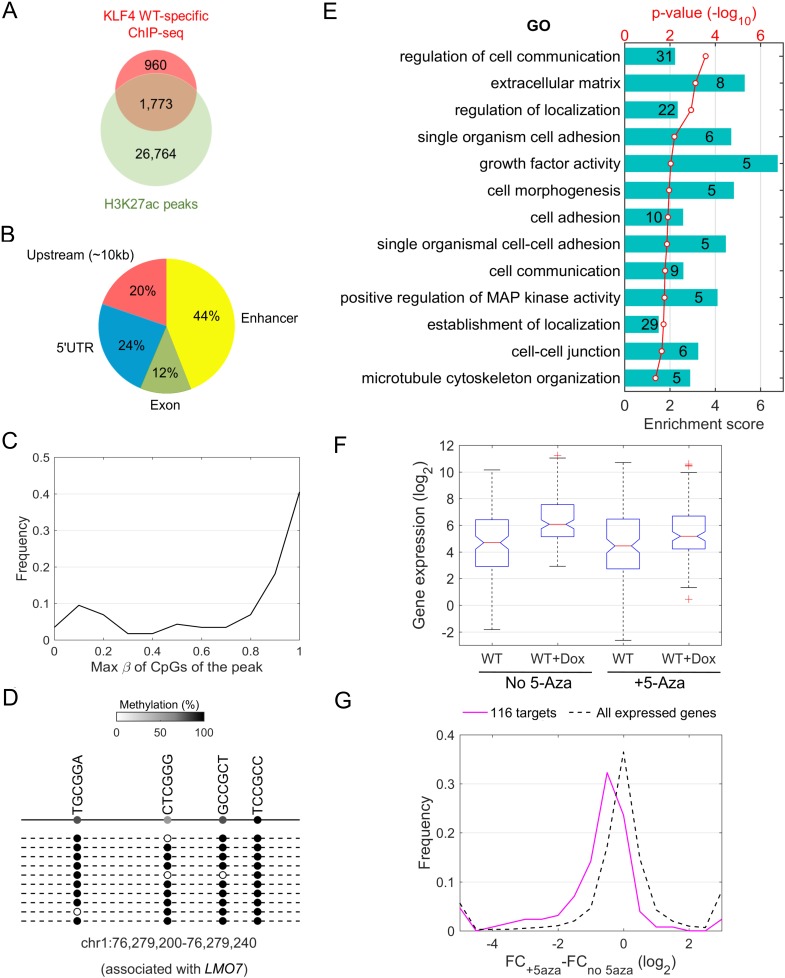
Figure 4. Downstream targets of KLF4-mCpG interactions.
(A) Overlap between KLF4 WT-specific ChIP peaks (2733) and the enhancer mark H3K27ac ChIP-seq peaks. A total of 1733 loci were identified. (B) A total of 116 KLF4-mCpG direct targets were identified in a serial of genome-wide studies. The overlap between WT-specific binding peaks (2733) and the 308 WT-only upregulated genes indicated that 20%, 24% and 12% of these genes were activated by KLF4 binding to mCpGs in gene upstream, 5’UTR and exon region, respectively. The overlap between enhancer regions in KLF4 WT-binding sites further identified 44% genes were activated by KLF4 binding to mCpGs in the enhancer regions. (C) Methylation level distribution of cis-elements in KLF4 binding peaks associated with 116 target genes. (D) Bisulfite sequencing confirmed DNA methylation in some of the KLF4 WT-specific binding peaks. (E) Gene ontology analysis of direct targets of KLF4-mCpG indicated that these targets have been implicated in cell adhesion, migration, cytoskeleton arrangement and cell binding activities. (F) Boxplot of gene expression for WT, WT+Dox, WT + 5 Aza, and WT + 5-Aza+Dox (G) Histogram of FC (after and before Dox) difference with and without 5-Aza for 116 target genes and all expressed genes, respectively.