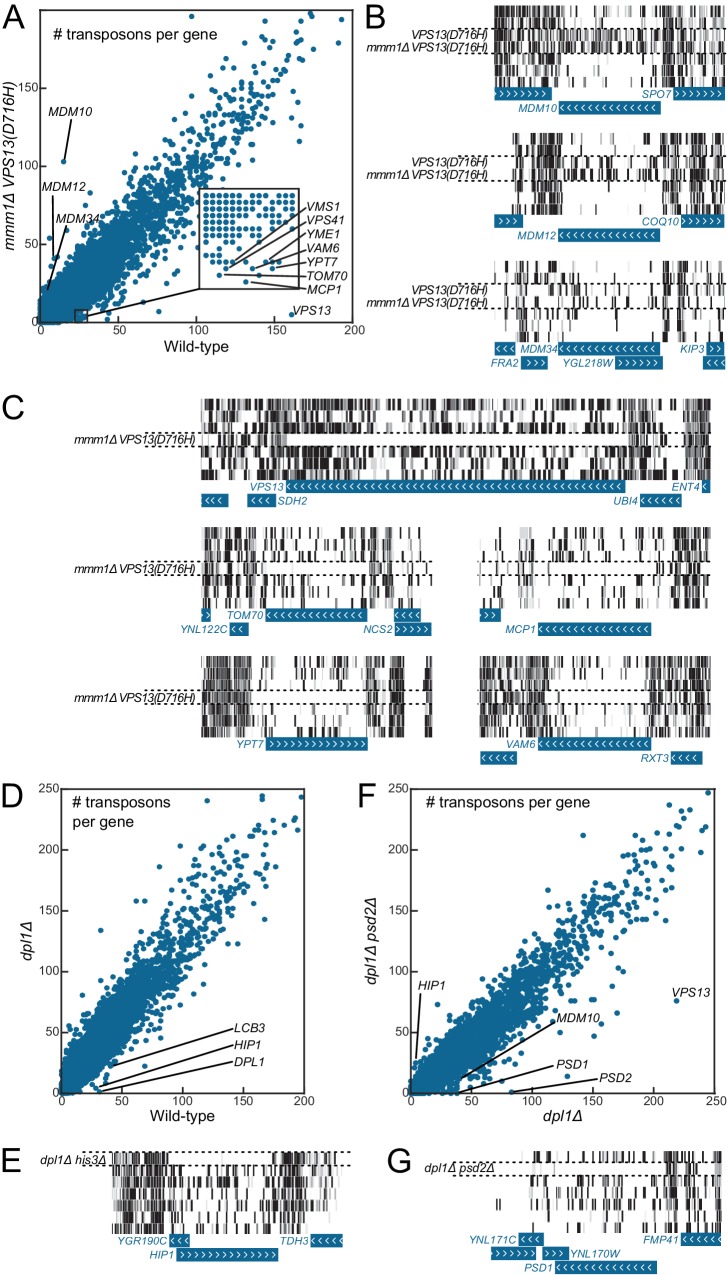
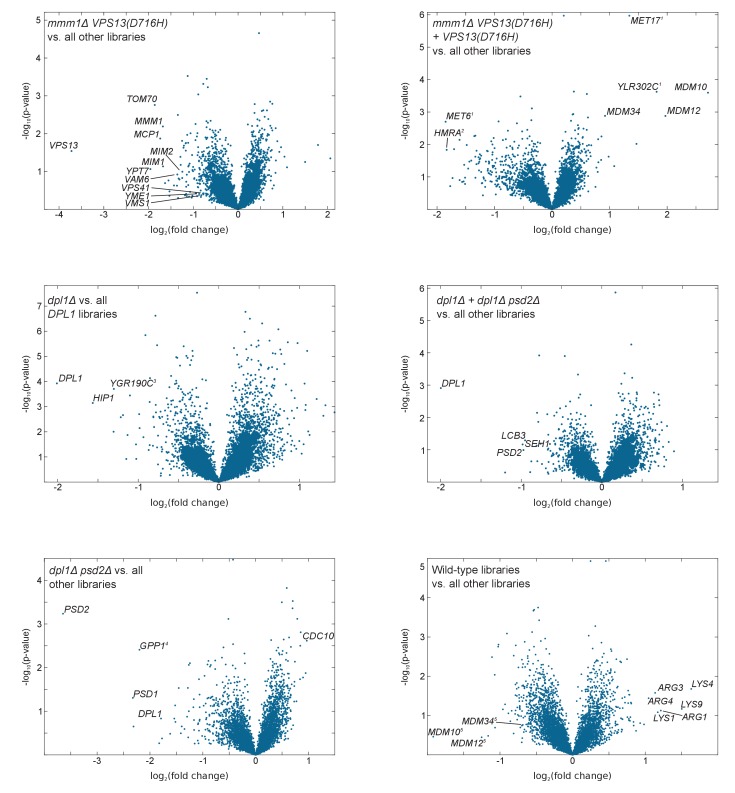
Figure 4. Genetic interaction analyses.
Libraries in panels B, C, E and G are displayed in the same order as in Figure 1C. (A) Comparison of the number of transposons inserted in each of the 6603 yeast CDSs in the wild-type (x-axis) and mmm1Δ VPS13(D716H) (y-axis) libraries. (B) Transposon coverage of genes encoding ERMES components is increased in libraries from strains bearing the VPS13(D716H) allele. (C) Examples of genes showing synthetic sick/lethal interaction with mmm1Δ VPS13(D716H). (D) Comparison of the number of transposons inserted in each of the 6603 yeast CDSs in the wild-type (x-axis) and dpl1Δ (y-axis) libraries. (E) Transposon coverage of the HIP1 locus in the dpl1Δ his3Δ library and in all the other libraries (HIS3). (F) Comparison of the number of transposons inserted in each of the 6603 yeast CDSs in the dpl1Δ (x-axis) and dpl1Δ psd2Δ (y-axis) libraries. (G) Transposon coverage of the PSD1 locus in the dpl1Δ psd2Δ and in all other libraries.