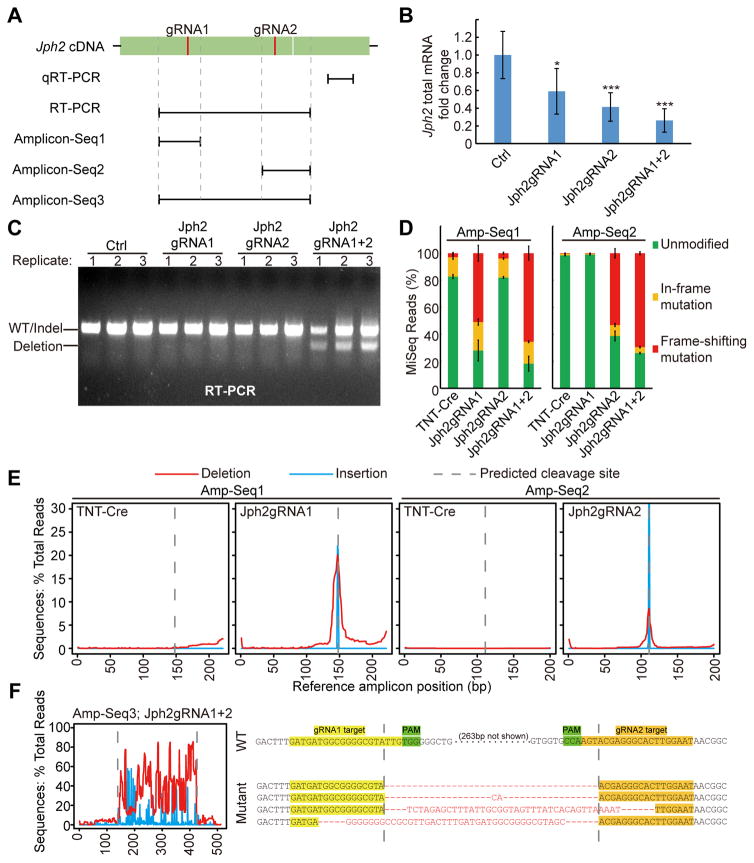
Figure 2. Multiple mechanisms mediate efficient Jph2 depletion in CMs.
(A) A scheme of targeted loci on Jph2 cDNA for qRT-PCR, RT-PCR and Amplicon-Seq analyses. (B) qRT-PCR analysis of Jph2 mRNA level. n=6 P7 heart apexes. Student’s t-test: *p<0.05; **p<0.01; ***p<0.001. (C) Expression of tandem gRNAs induced deletion between the gRNAs. Transcript structure was interrogated by RTPCR. (D) Quantification of in-frame and frame-shifting mutations identified by Amplicon-Seq. n=3 biological replicates. 300~2000 reads were analyzed per sample. (E) Distribution of small insertions and deletions along cDNA amplicon 1 and 2. (F) Analysis of mutation distribution in Amplicon 3 (left) reveals large deletions between gRNA1&2 target sites. Representative sequencing results in amplicon 3 are shown (right). Deleted or unaligned sequences are in red. Plots show mean ± SD in (B) and (D).