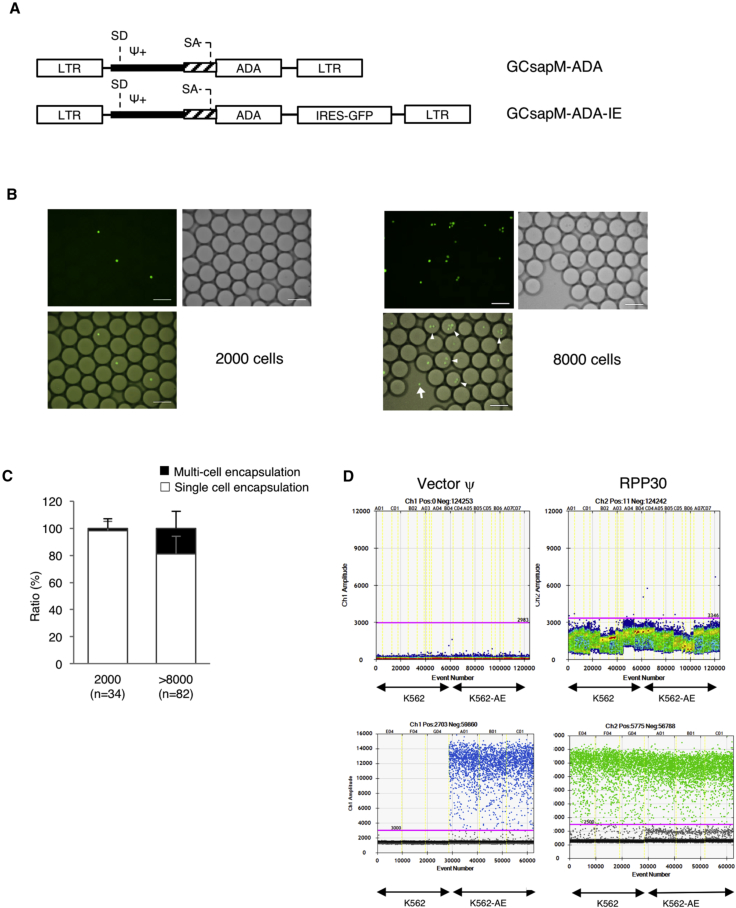
Figure 2.
Single-Cell Encapsulation and Direct PCR
(A) The structure of the retroviral vector used in this study. GCsapM-ADA was used in the clinical trials. EGFP cDNA was incorporated downstream of ADA cDNA with the sequence of an internal ribosomal entry site (IRES) in the pGCsapM-ADA-IE construct. K562 cells were transduced with GCsapM-ADA-IE to create K562-AE cells. (B) Direct encapsulation of K562-AE cells into droplets. The 2,000-cell samples exhibited single-cell encapsulation into droplets. Multi-cell encapsulation (triangles) and the failure of encapsulation (arrows) were observed in samples containing more than 8,000 cells. Fluorescent (EGFP) and bright-field (BF) images are shown. Scale bars represent 100 μm. (C) Ratios of droplets with single- or multi-cell encapsulation. Droplets were counted in 34 and 82 different fields on microscopy in the 2,000- and > 8,000-cell samples, respectively. (D) Direct PCR subsequent to single-cell encapsulation. Droplets containing a single cell displayed fluorescent signals after amplification of the vector ψ and reference gene RPP30. Upper panels, standard procedure; lower panels, modified procedure. LTR, long terminal repeat; SA, splice acceptor; SD, splice donor.