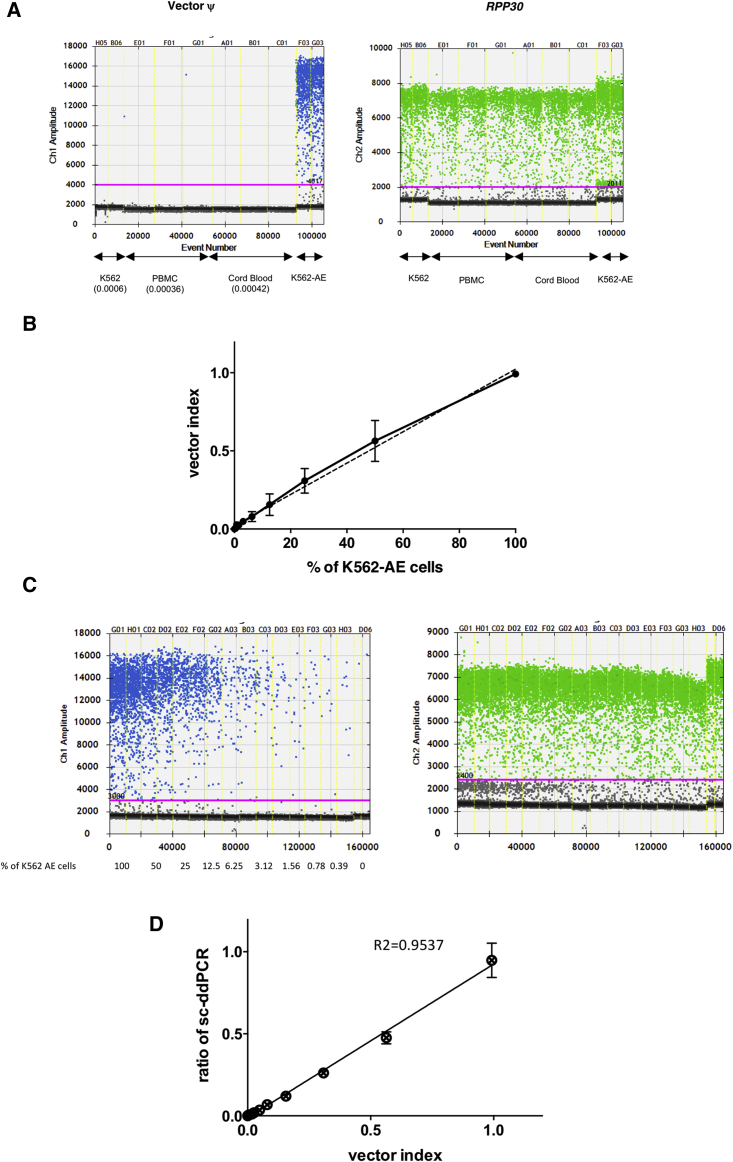
Figure 3.
Estimation of the Accuracy of Single Cell-Based Digital Droplet PCR
(A) Evaluation of the non-specific signals in negative samples. Target ψ and RPP30 were amplified in mononuclear cell samples of peripheral blood (PBMCs) and cord blood from healthy donors, as well as naive K562 cells. The ratio of the target ψ, which denotes the background signal, is shown below each sample. (B) Relationship between the percentages of dilution and the vector index in extracted genomic DNA from spiked cell samples. K562 cell samples were spiked with serially diluted K562-AE cells carrying the vector at a concentration of one copy per cell. Vector ψ and RPP30 were measured using genomic DNA from spiked samples by conventional ddPCR. The vector index was calculated using the following formula: (2 × number of vector-positive droplets)/(numbers of RPP30-positive droplets). An index of 1 indicates that all cells contain one copy of ψ and two copies of RPP30 in their genomes. The measured value in each spiked sample was linearly related to the theoretical values. (C) Single cell-based digital droplet PCR (sc-ddPCR) using spiked samples. K562 cell samples spiked with serially diluted K562-AE cells were analyzed by sc-ddPCR. The number of ψ signal-positive droplets, which contain vector-positive cells, declined in relationship with the spiked ratios, whereas similar numbers of RPP30-positive droplets were observed irrespective of the spiked ratio, which indicated the sample size. (D) Correlation between the vector index and the ratio of vector-positive droplets as determined by sc-ddPCR. sc-ddPCR was performed using spiked K562 cells, and the measured values were plotted against the vector index in genomic DNA. The ratios determined by sc-ddPCR were linearly related to the vector index.