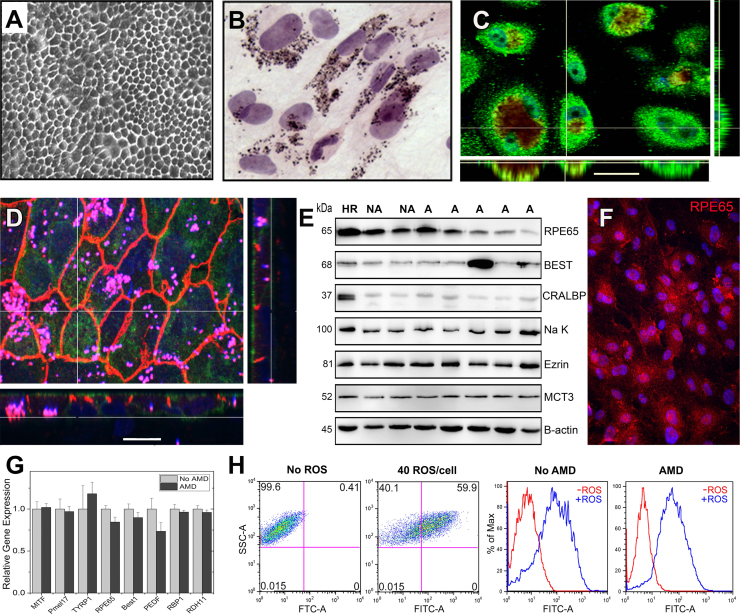
Fig. 1.
Characterization of primary human RPE cultures derived from adult donors. (A) Phase microscopy image shows confluent RPE form a monolayer with a cobblestone appearance. (B) H&E stained RPE cells visualize the pigment granules inside the cytoplasm (C,D) Confocal microscopy images of RPE cultured on transwell inserts for one month are shown. En face views of the RPE monolayer shown as maximum intensity projections through the z-axis. Also shown are cross-sections (locations shown by the white line) through the z-plane of multiple optical slices. (C) Bestrophin (green) is expressed on the basal surface. Pigment granules seen due to auto fluorescence (red) Nuclei are stained using DAPI (blue). (Scale bar 30 µm) (D) The Na-K ATPase (green) is expressed on the apical surface. ZO-1 staining (orange) marks cell borders; punctate pigment granules appear pink. Nuclei are stained using DAPI (blue). (Scale bar 15 µm) (E) RPE cultures from non-diseased (No AMD) and AMD donors contain many prototypic RPE proteins as demonstrated on Western immunoblots. Molecular mass for each protein is shown on the left. HR is RPE homogenate from a human donor. β-actin is the loading control. CRALBP, cellular retinaldehyde-binding protein; MCT3, monocarboxylate transporter 3. (F) Immunohistochemistry and fluorescent microcopy was used to detect the expression of the prototypic RPE protein, the 65 kDa Retinal Pigment Epithelium-Specific Protein RPE65. (G) RT-qPCR analysis was performed on RPE cultures from non-diseased (No AMD) (n=4) and AMD (n=7) donors. Graph shows AMD delta CT relative to the mean delta CT of the non-diseased group. Data are the mean (±SEM) normalized values. MITF, microphthalmia-associated transcription factor; Pmel17, pre-melanosome protein 1; TYRP1, tyrosinase related protein 1; Best1, Bestrophin; PEDF, pigment epithelial derived factor; RBP1,Retinol binding protein 1; RDH11, Retinal dehydrogenase. (H) FACs analysis measuring the phagocytosis of FITC-labeled rod OS by RPE from a healthy (No AMD) and an AMD donor. Dot plots (left) and histograms for cells without and with the addition of OS are shown. Data are mean (± SEM). (* denotes p<0.05).