Fig. 2.
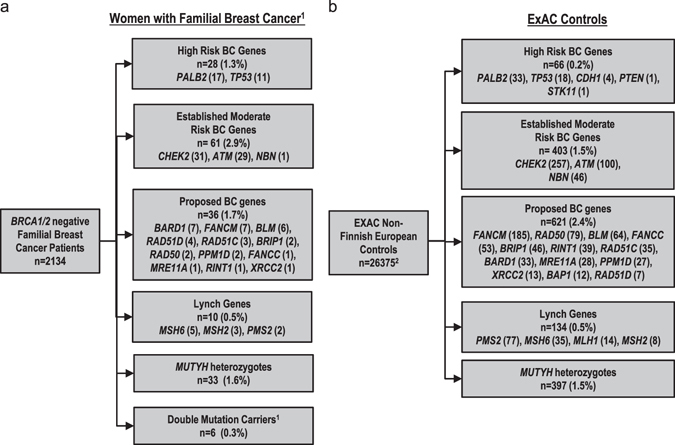
Spectrum of mutations in 26 genes in familial breast cancer women compared to Exome Aggregation Consortium (ExAC) controls. Rates of truncating, known pathogenic missense mutations, and copy number variants in (a) BRCA1/2 negative familial breast cancer women analyzed by targeted sequencing and (b) analysis of ExAC data (excluded copy number variation). Sequencing data was analyzed for mutations in high risk breast cancer genes, moderate risk breast cancer genes, proposed breast cancer genes, the Lynch syndrome genes, and MUTYH. 1Women with two pathogenic mutations were removed from single gene counts and include two cases with ATM/CHEK2 mutations, two with CHEK2/PALB2 mutations, and one patient each with ATM/RAD50, and MSH6/RAD50 mutations. 2Denominator for ExAC non-Finnish European controls (The Cancer Genome Atlas excluded) was determined by averaging the allele number for each identified variant and dividing by two.
