Figure 5.
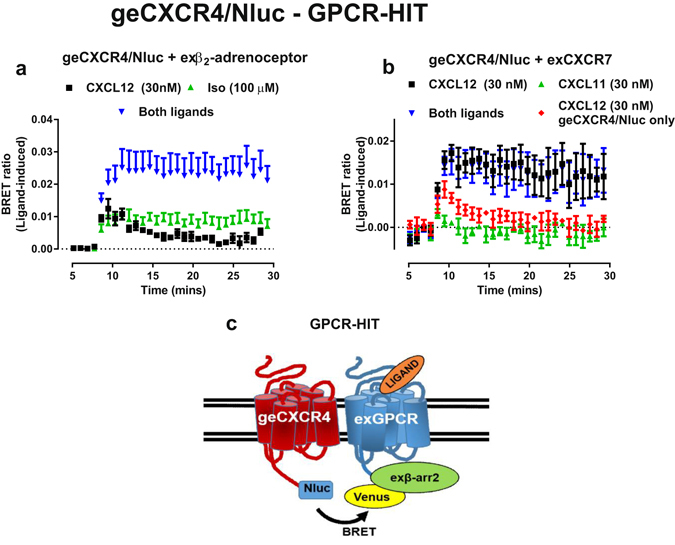
Demonstration of GPCR-HIT BRET assay using genome-edited CXCR4. HEK293FT cells expressing genome-edited CXCR4 fused to Nluc (geCXCR4/Nluc) transiently transfected with cDNA coding for β-arrestin2/Venus and (a) β2-adrenoceptors or (b) CXCR7 were used to carry out BRET assays using the GPCR-HIT configuration. (a) Cells were stimulated with CXCL12 (30 nM, black squares), isoprenaline (100 µM, green upward triangles) or both ligands simultaneously (blue downward triangles) or (b) CXCL12 (30 nM, black squares), the CXCR7-specific ligand CXCL11 (30 nM, green upward triangles) or both ligands simultaneously (blue downward triangles). (b) For comparison, responses mediated by CXCL12 (30 nM, red diamonds) in HEK293FT cells expressing genome-edited CXCR4 fused to Nluc (geCXCR4/Nluc) transiently transfected with only cDNA coding for β-arrestin2/Venus is shown. (c) Schematic representation of the GPCR-HIT configuration using NanoBRET. ‘BRET ratio (ligand-induced)’ was calculated as described in Methods. Points represent mean BRET ratio ± S.E.M.; bars represent maximum corrected BRET ratio ± S.E.M. observed in kinetic time courses. Ligand added at approximately 8 minutes. Data generated from three independent experiments.
