Fig. 1.
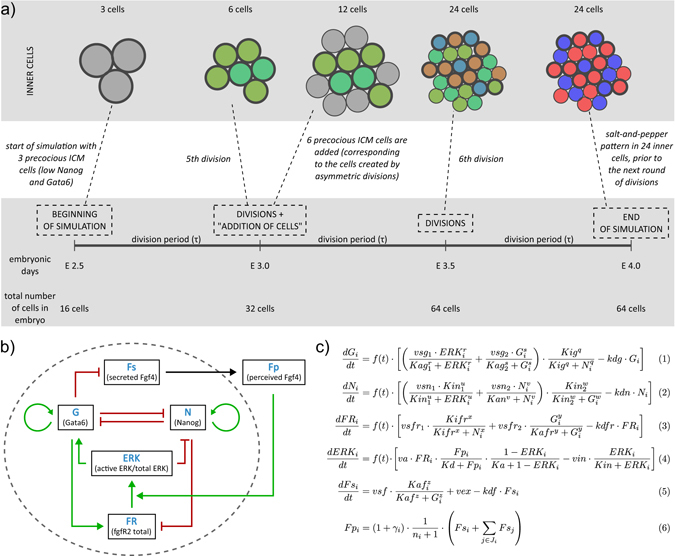
a Scheme of the model of development of epiblast (Epi) and primitive endoderm (PrE) lineage from inner cells (ICM). The model does not consider trophectoderm explicitly and starts from three inner cells originating from the first round of differentiative divisions (In1). The three cells begin the process of specification to Epi and PrE cells. In the second round of divisions, the three In1 cells divide into six cells, whereas differentiative divisions add six ICM cells (In2). Thus, at E3.0 the inner part of the embryo consists of six In1 and six In2 cells. The development of Epi and PrE lineage and the emergence of the salt-and-pepper pattern continues until E4.0. The colour code is: grey for blastomers in which Nanog and Gata6 are low, green for ICM cells, red for Epi cells and blue for PrE cells. b Gene regulatory network (GRN) present in each of the modelled cells. c Equations of the GRN model, describing the rate of change of concentrations of Gata6 (G), Nanog (N), FgfR2 (FR), secreted Fgf4 (Fs) and perceived Fgf4 (Fp) as well as the level of activity of the Erk pathway (ERK). Index i denotes the ith cell and n i the number of neighbouring cells. See text for details. Definitions and values of parameters used in simulations can be found in Supplementary Tables S1–S3
