Fig. 9.
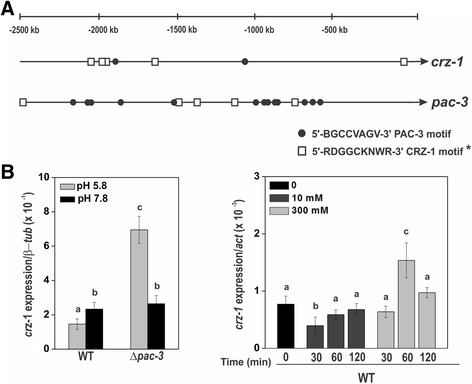
The influence of pH and Ca2+ on the crz-1 expression. a Schematic representation of the crz-1 and pac-1 promoters and the respective transcription factors DNA binding sites. The black dots and square boxes indicate the position of PAC-3 and CRZ-1 motifs, respectively. b Expression of crz-1 gene in Δpac-3 and wild-type strains under normal and alkaline pH (left graph) and under different CaCl2 concentration (right graph). Cells from both strains were cultured at pH 5.8 during 24 h and shifted to pH 7.8 during 1 h. Cells from the wild-type strain were cultured at normal pH (0) for 24 h and shifted to 10 or 300 mM CaCl2 for 30, 60 and 120 min. Mycelial samples were collected and used to extract total RNA. Gene expression analysis was performed by RT-qPCR in the StepOnePlus™ Real-Time PCR system (Applied Biosystems) using the Power SYBR® Green and specific primers (Additional file 1: Table S1, qPCR). The tub-2 and act genes were used as the reference genes in the pH and calcium experiments, respectively. At least three biological replicates were run and the data were analyzed in the relative quantification standard curve method. a, b, c: Letters above bars indicate statistical significance; different letters indicate significant differences between two samples and equal letters indicate no significant difference between two samples in the same or different pH (Student’s t-test, P < 0.01). * N. crassa DNA binding site according to Weirauch et al. [33]
