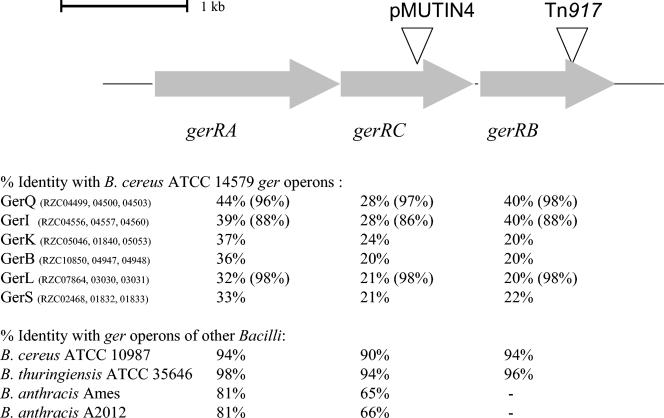
FIG. 1.
Organization of the gerR operon and homology of the gerR operon with ger operons of B. cereus ATCC 14579 and other bacilli. Triangles show the positions of Tn917 insertion for mutant strain LH104 and of pMUTIN4 insertion for mutant strain LH129. The code numbers of the open reading frames of the gerR operon in the B. cereus ATCC 14579 genome database (10) are RZC04487 (A), RZC04488 (C), and RZC04490 (B). The code numbers of the open reading frames of the other ger operons and their percentages of amino acid identity are indicated. In contrast to gerR, these ger operons have the gene order gerA gerB gerC. The gene products encoded by the gerQ, gerI, and gerL operons are described for B. cereus 569 (1, 2), and the percentages of homology of these operons with the gerQ, gerI, and gerL operons of B. cereus ATCC 14579 are shown between brackets. Close homologues of the gerR operon can be found in other bacilli. The percentages of amino acid identity with homologues of the gerR operon in B. cereus ATCC 10987 (21), B. thuringiensis subsp. israelensis ATCC 35646 (http://www.ergo-light.com), B. anthracis Ames (http://www.tigr.org), and B. anthracis A2012 (http://www.ergo-light.com), are indicated.