Abstract
Temporal and spatial dynamics of ammonia-oxidizing bacteria (AOB) were examined using genes encoding 16S rRNA and ammonia monooxygenase subunit A (AmoA) in Monterey Bay, Calif. Samples were collected from three depths in the water column on four dates at one mid-bay station. Diversity estimators for the two genes showed a strong positive correlation, indicating that overlapping bacterial populations had been sampled by both sets of clone libraries. Some samples that were separated by only 15 m in depth had less genetic similarity than samples that were collected from the same depth months apart. Clone libraries from the Monterey Bay AOB community were dominated by Nitrosospira-like sequences and clearly differentiated from the adjacent AOB community in Elkhorn Slough. Many Monterey Bay clones clustered with previously identified 16S rRNA and amoA groups composed entirely of marine sequences, supporting the hypothesis that these groups are specific to the marine environment and are dominant marine AOB. In addition, novel, phylogenetically distinct groups of AOB sequences were identified and compared to sequences in the database. Only one cluster of gammaproteobacterial AOB was detected using 16S rRNA genes. Although significant genetic variation was detected in AOB populations from both vertical and temporal samples, no significant correlation was detected between diversity and environmental variables or the rate of nitrification.
Aerobic nitrification is the microbially mediated two-step oxidation of ammonia to nitrite and then to nitrate. The two steps are performed by different groups of bacteria, the ammonia-oxidizing bacteria (AOB) and the nitrite oxidizers. The process of nitrification is important in aquatic environments, because it links N mineralization, the formation of ammonia from organic materials, to denitrification, which results in the loss of nitrogen from the system in the form of gaseous dinitrogen (43). This linkage is especially important in coastal aquatic systems where anthropogenic inputs of nitrogen can result in heavy nutrient loading, and denitrification can account for the loss of up to 50% of the inorganic N input (34). Disruption of nitrification, denitrification, or the linkage of these processes can lead to eutrophication and system degradation.
The evolutionary history and genetic diversity of AOB have been examined in numerous environments with AOB-specific 16S rRNA primers (for examples see references 2, 7, 14, 20, 36, and 39). Unlike most functional groups, AOB-specific 16S rRNA amplification is feasible because the AOB are restricted to two monophyletic clusters within the beta- and gamma-Proteobacteria (9, 28).
A second gene that can be used to investigate AOB diversity is amoA, which encodes the first subunit of ammonia monooxygenase, the protein that catalyzes the oxidation of ammonia to hydroxylamine (21), which is then converted to nitrite (11). amoA has two major advantages over 16S rRNA for comparisons of genetic diversity. First, amoA encodes a protein involved directly in ammonia oxidation, therefore genetic differences are more likely to be of functional importance to the process of interest, nitrification. Second, the rate of molecular divergence in amoA is expected to exceed that of 16S rRNA (28), allowing for greater resolution of genetic differences in natural populations. Unlike the evolutionary history of many functional genes, horizontal transfer of amoA in AOB appears to be minimal or absent. This provides a parallel phylogenetic topology for amoA and the gene encoding 16S rRNA, which simplifies inference of the evolutionary history of the organism based upon these genes.
Although the number of studies is small, common insights are beginning to emerge about the diversity and composition of marine AOB assemblages. For example, the dominant betaproteobacterial AOB 16S rRNA gene sequence in clone libraries from the polar oceans (2, 10) has also been observed as a dominant sequence in clone libraries from the Mediterranean Sea (27). As sequence data sets represented as phylogenetic trees increase in size, visual interpretation of patterns is no longer adequate to examine divergence within and between samples. The quantitative genetic comparisons utilized in this study help to interpret patterns of diversity and allow quantitative comparisons to environmental variables and biogeochemical function, such as nitrification rate. This type of analysis will be vital in the interpretation of the large sequence data sets required for the analysis of system dynamics, an important step in the transition to true molecular ecology of microbial systems.
The present study of AOB at a mid-latitude Pacific site had three main goals: (i) to elucidate the dynamic diversity and structuring of AOB communities using both amoA and 16S rRNA genes from one station at three depths and four dates in Monterey Bay (MB), Calif.; (ii) to compare AOB communities in MB and the adjacent estuarine system of Elkhorn Slough in an attempt to differentiate regional supply from biological selection in neighboring, but sharply contrasting, ecosystems; and (iii) to investigate potential links between environmental variation, AOB diversity, and biogeochemical function by utilizing pairwise comparisons of AOB diversity, nitrification rates, and measured hydrographic, nutrient, and biological variation.
MATERIALS AND METHODS
Sampling.
Bimonthly sampling cruises began in February 1998 and ended in October 1999. Water samples were collected throughout the euphotic zone (defined as the 0.1% light level) at a mid-bay station (depth, 900 m; 36°45′N, 122°01′W) in Monterey Bay, Calif., using a 10-liter Niskin rosette mounted on a conductivity-temperature-depth (CTD) sensor. This sampling scheme was used to cover the range of nitrification rates in the water column, because the nitrification maximum typically occurs above the depth of 0.1% light but below the depth of 10% light (43). Temperature and salinity measurements (Table 1) were made using a Seabird CTD sensor, and subsamples were collected for nutrient analysis. A complete analysis of the temporal variability in hydrographic and biogeochemical parameters from the 2-year period is under way (44). Six samples were analyzed for molecular diversity of AOB communities: April 98-M (M, mid-depth within photic zone), October 98-M, April 99-M, October 99-S (S, surface depth; 93% light), October 99-M, and October 99-D (D, deep; 0.1% light) (Table 2). Seawater (4 liters) was concentrated using tangential flow filter cassettes (Ultrasette 300 KD open channel; Filtron Technology Corp., Northborough, Mass.). The retentate was collected on 0.2-μm-pore-size supor filters (Gelman), frozen immediately, and archived at −80°C until DNA was extracted.
TABLE 1.
Environmental parameters and biogeochemical rates for samples used for sequence analysis
| Sample | Depth (m) | No. of betaproteobacterial amoA clones sequenced (450 bp) | No. of betaproteobacterial 16S rRNA clones sequenced (295 bp) | Temp (°C) | Nitrite (nM) | Nitrate (μM) | Bacteria (105 cells/ml) | Nitrification (nM per day) |
|---|---|---|---|---|---|---|---|---|
| 4/98-M | 40 | 11 | 9 | 9.5 | 9 | 29.05 | 5.41 | 68.6 |
| 10/98-M | 32 | 11 | 10 | 13.0 | 68 | 5.50 | 5.69 | 73.8 |
| 4/99-M | 33 | 8 | 9 | 9.5 | 18 | 11.41 | 3.37 | 27.8 |
| 10/99-S | 7 | 20 | 10 | 13.0 | 15 | 8.80 | 10.40 | 28.0 |
| 10/99-M | 36 | 21 | 9 | 11.7 | 30 | 18.30 | 5.53 | 38.2 |
| 10/99-D | 50 | 20 | 10 | 11.2 | 10 | 15.70 | 3.94 | 52.0 |
TABLE 2.
Integrated nitrification rates (integrated over the 100 to 0.1% light depth)
| Date | Integrated nitrification rate (nmol m−2 day−1) | Depth (m) of 1% light | Depth (m) of nitrite maximum (conc. [nM]) |
|---|---|---|---|
| April 1998 | 2.29 | 36.5 | 43 (15) |
| October 1998 | 3.30 | 48.6 | 32 (68) |
| April 1999 | 1.85 | 48.6 | 33 (46) |
| October 1999 | 1.38 | 28.4 | 23 (210) |
Biogeochemical measurements.
Nitrate and nitrite concentrations were measured using colorimetric techniques, either manually (37) or using a Lachat autoanalyzer. The nitrification rate was measured with the 15N tracer method of Ward (41) (Tables 1 and 2) using 4-liter, 4- to 6-h incubations under simulated in situ conditions in deck incubators.
DNA extraction and gene amplification.
DNA was extracted from the filters with a phenol-chloroform extraction (1) and were resuspended in 100 μl of sterile water. DNA (1 μl) from each extraction was amplified with universal bacterial primers EUB 1 and 2 (Escherichia coli positions 9 to 1531) (15) to test the PCR quality of DNA extracts. Using 1 μl of the EUB amplification product as the template, AOB 16S rRNA gene fragments were amplified with beta (NIT A,B; ∼1,080 bp)- and gamma (NOC 1,2; ∼1,100 bp)-proteobacterial AOB-specific primers and cycling parameters (39, 46). The amoA gene fragments (∼490 bp) were amplified from DNA extracts using the PCR primers (AmoA-1F and AmoA-2R) and cycling parameters of Rotthauwe et al. (30), modified with an annealing temperature of 55°C. This amoA primer set is specific to the betaproteobacterial AOB and does not amplify amoA gene fragments from gammaproteobacterial AOB. PCR products were separated by electrophoresis in 1% agarose gels. Appropriately sized fragments were cloned using the TOPO-TA cloning kit (Invitrogen) and were screened directly by PCR for the presence of inserts using T7 and M13R vector primers.
Sequencing and phylogenetic analysis.
Sequencing reaction mixtures from the T7/M13R PCR products were prepared using Big Dye terminator chemistry (version 3.1; Perkin-Elmer) and run on either an ABI 3100 or an ABI 310 capillary sequencer (PE Applied Biosystems). Sequence reaction mixtures for NIT A,B and NOC 1,2 fragments were also prepared with internal universal 16S rRNA primers DS907R and GM5F (38) to obtain overlapping coverage of the entire fragment for some clones. The majority of NIT clones were only partially sequenced (295 bp). Double coverage of this 295-bp NIT sequence was obtained by duplicate sequence reactions from the NIT A end of the fragment (starting at position 165 of E. coli 16S rRNA) with either the T7 or M13R primer, depending on the orientation of the cloned fragment within the vector. The number of clones analyzed from each depth is shown in Table 1. Sequencher (GeneCodes) software was used to generate nucleotide sequence alignments. AmoA amino acid alignments were created using the Clustal X software package (version 1.81) (12). The constructed alignments were analyzed with the PAUP software package (version 4.0b10). Neighbor-joining phylogenetic trees were created from Jukes-and-Cantor-corrected distances and 1,000 bootstrap replicates.
Statistical analysis of sequences.
The possibility of PCR bias causing deviations in clonal abundance and composition must be acknowledged and is unavoidable (17). Thus, it is emphasized that the following comparisons must be viewed as applying to the clone libraries per se rather than to the original samples. All statistical analyses of the betaproteobacterial AOB 16S rRNA were performed with the partial-length (295 bp) fragment. The levels of genetic variation within and between samples were calculated with the Arlequin population genetics software package (32). The partitioning of variation within and among samples was estimated with an analysis of molecular variance (AMOVA) (5). Using sequence data, the mean number of pairwise genetic differences (π) (23) within (πw) and between (πB) samples was calculated. Pairwise dissimilarity indices (Fst) (19, 29) were calculated for pairs of samples as follows: Fst = (πB) − (average of πw for the two samples)/(πB). The possible values of Fst range from 1 (if all of the variation occurs between samples) to 0 (if the variation within samples is equal to the variation between samples). The statistical significance of nonrandom distributions was tested with 100 permutations in which random assemblages of the sequences were compared to observed distributions. Correlation analyses were performed with the JMP statistical package (31).
Nucleotide sequence accession numbers.
The amoA sequences reported in this study have been deposited in GenBank under accession numbers AY736856 to AY736946. The betaproteobacterial and gammaproteobacterial 16S rRNA sequences in this study have been deposited in GenBank under accession numbers AY736947 to AY737003 and AY744690 to AY744700, respectively.
RESULTS AND DISCUSSION
Environmental parameters and nitrification rates.
Vertical profiles of temperature and nitrite concentration (Table 1) for the six samples analyzed in this study were consistent with those of a mixed-water column. Nitrification rates, integrated over the water column, did not display a clear seasonal pattern and varied by a factor of only 2.5 over the 2-year period (44). A similar range of variation was found among the four dates included in this study (Table 2). The nitrification rate maximum occurred at 40 to 60 m, and it ranged from 27 to 74 nmol per liter per day (Table 1). Surface samples (<20 m) had the lowest nitrification rates (21 to 28 nmol per liter per day).
Betaproteobacterial AOB 16S rRNA diversity.
An analysis of molecular variance of the betaproteobacterial AOB 16S rRNA MB clones indicated that 59.2% of the total variation occurred within the samples, versus 40.8% among samples (data not shown). Thus, 40.8% of the observed variation depended on comparisons between clone libraries and was not observed within a single library. Diversity estimators (πW and πB) and pairwise dissimilarity indices (Fst) were used to examine the partitioning of this variation within and between the samples. All values for the mean number of pairwise sequence differences within a sample (πW) were similar and small (<4.0), i.e., sequences within a sample were similar, except for the October 99-M sample (πW = 13.94) (Table 3). This suggests that the 16S rRNA sequences in the October 99-M clone library were not very similar to each other compared to levels of similarity within the other libraries. A similar pattern of variation was also found in comparisons between samples. All values for the mean number of pairwise differences between samples (πB) were similar and small (<4.0), except for pairwise comparisons to the October 99-M sample (πB = 17.21 to 18.12; Table 3). These πW and πB values indicate that, excluding the October 99-M sample, the gene sequences found within a clone library (from one sample) were similar, and when the sequences in two libraries (two samples) were compared, they were also very similar to one another. This is consistent with the relationship of samples in the 16S rRNA phylogenetic tree (Fig. 1A). Although levels of divergence with 16S rRNA were limited, Fst values, estimating the partitioning of variation within versus between samples, indicated a nonrandom structuring in all pairwise comparisons of samples except for the pairings of April 99-M with October 98-M (P = 0.06), October 98-M with October 99-D (P = 0.10), and October 99-S with October 99-D (P = 0.59). This indicates that although most libraries were similar, some variation was detected and stochastic sampling alone could not explain this variation (except in the pairs identified above); i.e., although most samples were similar, meaningful community structure was observed in time and space.
TABLE 3.
Diversity estimators for pairwise comparisons of 16S rRNA sequences
| Site | Sequence difference for site:
|
|||||
|---|---|---|---|---|---|---|
| 4/98-M | 10/98-M | 4/99-M | 10/99-S | 10/99-M | 10/99-D | |
| 4/98-M | 2.78a | 0.09c | 0.14 | 0.11 | 0.54 | 0.08 |
| 10/98-M | 3.46b | 3.53 | 0.10* | 0.07 | 0.53 | 0.03* |
| 4/99-M | 3.70 | 3.97 | 3.61 | 0.13 | 0.51 | 0.12 |
| 10/99-S | 2.85 | 3.12 | 3.40 | 2.28 | 0.53 | −0.02* |
| 10/99-M | 18.10 | 18.12 | 18.09 | 17.21 | 13.94 | 0.56 |
| 10/99-D | 2.78 | 3.05 | 3.36 | 2.28 | 17.48 | 2.36 |
Mean pairwise sequence differences within (πW) sites are on the diagonal in boldface.
Mean pairwise sequence differences between (πB) sites are below the diagonal in italices.
Pairwise population Fst values are above the diagonal. An asterisk indicates a nonsignificant (P > 0.05) observed Fst value compared to Fst values from 100 randomizations of the sequences; i.e., it indicates random structuring of sequences.
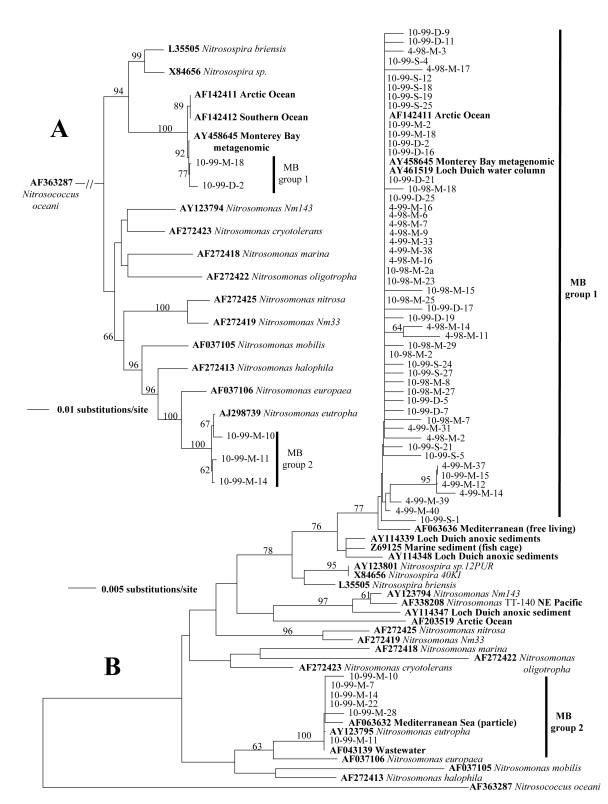
FIG. 1.
Distance neighbor-joining trees of the betaproteobacterial AOB 16S rRNA based on (A) a subset of cloned sequences based on the 1,060-bp fragment or (B) the 295-bp fragment. Database sequences for cultured strains and environmental sequences are in boldface and are identified by GenBank accession numbers. Monterey Bay sequences are labeled as month-year-depth (surface [S], mid [M], or deep [D])-clone number; e.g., 10-99-S-18 represents October 1999 surface clone 18. Bootstrap values of >50 are displayed.
Betaproteobacterial AOB amoA diversity.
AMOVA of the amoA MB clones indicated that 52.0% of the total variation occurred within the samples versus 48.0% between samples, suggesting that the sequences within individual clone libraries account for approximately half of the total observed variation. This is consistent with the patterns displayed on the phylogenetic tree shown in Fig. 2. The smallest πW for amoA was found in the sample from April 98-M (Table 4), while the largest πW was found within the October 99-M sample. The smallest pairwise πB indicates that samples October 99-D and April 98-M are most similar to each other. In contrast, the most divergent pair of samples is October 99-M and April 98-M. The largest pairwise differences (πB) in this study all involve comparisons to the October 99-M sample. Fst comparisons show a nonrandom structuring in all pairwise comparisons except for the pairing of April 99-M with October 98-M (P = 0.32). This indicates that both spatial and temporal structure was observed in the amoA sequence data set that was not due to stochastic sampling.
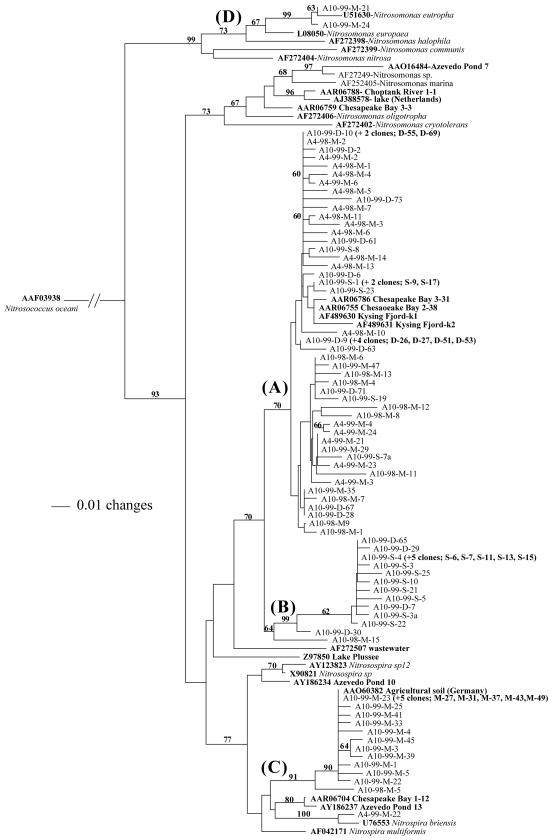
FIG. 2.
Distance neighbor-joining tree of betaproteobacterial AOB AmoA sequences based on 149 amino acid residues. Database sequences for cultured strains and environmental sequences are in boldface and are identified by GenBank accession numbers. Monterey Bay sequences are labeled A (for AmoA)-month-year-depth (surface [S], mid [M])-clone number; e.g., A10-99-M-21 represents AmoA October 1999 mid-depth clone 21. Bootstrap values of >60 are displayed.
TABLE 4.
Diversity estimators for pairwise comparisons of amoA sequences
| Site | Sequence difference for site:
|
|||||
|---|---|---|---|---|---|---|
| 4/98-M | 10/98-M | 4/99-M | 10/99-S | 10/99-M | 10/99-D | |
| 4/98-M | 10.91a | 0.49c | 0.34 | 0.54 | 0.67 | 0.14 |
| 10/98-M | 51.02b | 41.33 | 0.01* | 0.44 | 0.54 | 0.21 |
| 4/99-M | 42.11 | 44.90 | 47.89 | 0.41 | 0.52 | 0.12 |
| 10/99-S | 59.24 | 70.76 | 70.93 | 38.58 | 0.57 | 0.26 |
| 10/99-M | 111.84 | 106.72 | 107.38 | 108.72 | 53.85 | 0.56 |
| 10/99-D | 32.50 | 53.40 | 51.08 | 55.25 | 110.99 | 42.70 |
Mean pairwise sequence differences within (πW) sites are on the diagonal in boldface.
Mean pairwise sequence differences between (πB) sites are below the diagonal in italices.
Pairwise population Fst values are above the diagonal. An asterisk indicates a nonsignificant (P > 0.05) observed Fst value compared to Fst values from 100 randomizations of the sequences; i.e., it indicates random structuring of sequences.
Betaproteobacterial AOB 16S rRNA phylogenetic analysis.
Phylogenetic relationships of 16S rRNA sequences were investigated using a 295-bp fragment (Fig. 1A) for all 57 sequences from this study and using a 1,060-bp fragment (Fig. 1B) for a subset of five sequences (Table 1). The topology of the two distance-based neighbor-joining trees was compared to ascertain the degree to which the relationships were adequately represented by an analysis based on the smaller fragment. Bootstrap values of database clusters were higher with the larger fragment, but the clustering patterns in the two trees were consistent, so the following analyses were based upon the larger sequence data set from the smaller fragment. The phylogenetic analysis of the 295-bp sequences showed that most (89%) of the Monterey Bay 16S rRNA sequences (Fig. 1A) were Nitrosospira-like and clustered with sequences from uncultivated marine organisms into MB group 1 of the 16S rRNA tree. In the present study, only sequences from the October 99-M sample clustered outside of MB group 1 (Fig. 1A). The main MB cluster contains members from group 1 of Stephen et al. (36), and other members have been described from polar marine environments (2, 10), the Mediterranean Sea (27), and the Loch Duich marine water column (7). No cultured strains have been described from this cluster, although it appears to be dominant in many marine environments. For example, sequences from this cluster account for 86% of the AOB clones in Arctic Ocean samples (2) and correspond to the dominant free-living AOB sequences reported from the Mediterranean Sea (27). An unpublished database sequence from a Monterey Bay metagenomic library also clusters with group 1. This further supports the dominance of sequences from group 1, because amplification-independent techniques presumably were used to obtain this sequence from the environment.
The second group of 16S rRNA sequences comprised the remaining 11% of the betaproteobacterial AOB MB clones. These sequences were Nitrosomonas-like and were most similar to Nitrosomonas eutropha (>99%). All of the sequences from MB group 2 (Fig. 1A) were found in the October 99-M sample. This group of sequences also clustered tightly with the dominant particle-associated AOB sequence found in the Mediterranean Sea (27). Only the October 99-M sample had sequences that clustered into both MB groups 1 and 2. The MB 16S rRNA sequences reveal limited diversity but high similarity to assemblages of AOB sequences that have been found in other marine environments. Hollibaugh et al. (10) hypothesized that the 16S rRNA sequences retrieved from the polar oceans (polar sequences in group 1; Fig. 1A) may correspond to the dominant polar AOB and possibly a dominant global marine AOB. The occurrence of nearly identical sequences in high frequency in Monterey Bay as well as the Mediterranean Sea (27) and Loch Duich (7) suggests that this AOB type may indeed be a dominant marine AOB not limited to polar habitats.
Gammaproteobacterial AOB 16S rRNA phylogenetic analysis.
Using the NOC 1,2 primers, a 1,100-bp fragment of 16S rRNA was examined for gammaproteobacterial AOB diversity. Two of six samples (April 99-M and Oct 99-M) yielded a PCR product with NOC primers, and this study reports new sequences from the April 99-M sample. These sequences represent the second set of gammaproteobacterial AOB 16S rRNA sequences that have been examined from environmental samples. Ward and O'Mullan (46) reported gammaproteobacterial AOB 16S rRNA sequences from the east and west coasts of the United States but found limited phylogenetic diversity. All of the environmental sequences reported in the previous study clustered tightly with cultured gammaproteobacterial AOB strains. The sequences reported by Ward and O'Mullan (46) included environmental clones previously sequenced from the Oct 99-M sample. In the present study, similar phylogenetic diversity was detected (data not shown) and all of the sequences clustered tightly with the sequences reported previously (46). The limited diversity detected in both of these studies could be due to specificity of the primer set used for amplification, or it could reflect limited phylogenetic diversity of gammaproteobacterial AOB in these environments.
Nitrosomonas serotypes were more abundant than Nitrosococcus serotypes in marine environments (40), typically by two orders of magnitude, but Nitrosococcus serotypes in the Chesapeake Bay reached a maximum abundance of 4.2 × 105 cells per liter (as opposed to 1.7 × 106 cells per liter for Nitrosomonas serotypes). In Pacific northwest marine and estuarine sediments, gammaproteobacterial AOB contributed minimally to amoA clone libraries compared to betaproteobacterial AOB (25). Ward and O'Mullan (46) reported limited molecular diversity but widespread distribution of genes encoding gammaproteobacterial AOB 16S rRNA in marine samples. The relative importance of gammaproteobacterial subdivision AOB in marine nitrification and the diversity of the betaproteobacterial versus gammaproteobacterial AOBs remain unclear but are important questions demanding further investigation.
Betaproteobacterial AOB AmoA phylogenetic analysis.
A distance-based neighbor-joining tree of 149 amino acid residues from AmoA was constructed (Fig. 2) with the 91 sequences from MB clones, together with relevant database sequences. AmoA displayed a more complex phylogenetic topology than 16S rRNA sequences, comprising four major groups (A, B, C, and D). Most sequences (58%) fell into group A and clustered with sequences from mesohaline and polyhaline Chesapeake Bay sediments (6) as well as with environmental clones from the sediments of Kysing Fjord (24), but they were distinct from any other published AOB sequences. Within group A, amino acid distances of >6% occurred between sequences, but these sequences did not form strongly supported subclusters, nor did they cluster with database sequences that could help to differentiate the subgroups. Group A contained sequences from every sample in this study, although the majority of October 99-M and October 99-S sequences were found outside of group A (in groups B and C, respectively). Group B represented 20% of the sequences examined and was composed primarily of October 99-S sequences plus three October 99-D sequences. The sequences in group B are <96% identical to group A sequences, but groups A and B are joined by a bootstrap value of 68 and are more closely related to each other than to any cultured strains.
Group C represents 20% of the sequences from the clone libraries and is composed primarily of sequences from the October 99-M sample. These sequences cluster with AmoA sequences from cultured Nitrosospira strains, brackish water sediment samples from the northern Chesapeake Bay (6), and sediment samples from Azevedo Pond, a site in Elkhorn Slough, Calif. (3). Groups A, B, and C (Fig. 2) may correspond to the 16S rRNA sequences in Nitrosospira-like group 1 (Fig. 1A), but this relationship of amoA and 16S rRNA environmental clones cannot be definitively tested with this data set. Groups A, B, and C are joined to Nitrosospira-like AmoA sequences with a bootstrap value of 60. The final group (D) contained only two sequences from the MB clone libraries, both from October 99-M, that cluster tightly with N. eutropha (>98% identity). This group is particularly interesting, because the only 16S rRNA Nitrosomonas-like sequences were also from the October 99-M sample and grouped tightly with N. eutropha. The divergence of the October 99-M sequences cannot be clearly attributed to any of the measured environmental variables (Table 1) (discussed below).
Comparison of 16S and amoA gene diversity.
Both 16S rRNA and amoA genes displayed similar patterns of partitioning of variation within versus between samples (see AMOVA results discussed above), indicating significant variation that is only detected in comparisons between samples. The greatest amount of variation for both genes was found in the October 99-M sample and similar patterns of variation between samples (πB), although different levels were observed for both genes. Fst values for 16S rRNA sequences indicated a slightly larger number (3 of 15; Table 3) of sample pairs with randomly distributed sequences than amoA Fst values (1 of 15; Table 4), but the greater resolution (relative amount of sequence divergence) of amoA would allow samples to be more easily differentiated. This analysis also reveals that samples separated by <15 m at the same station can have higher levels of genetic differentiation than samples that are temporally separated in distinct seasons or years (Tables 3 and 4).
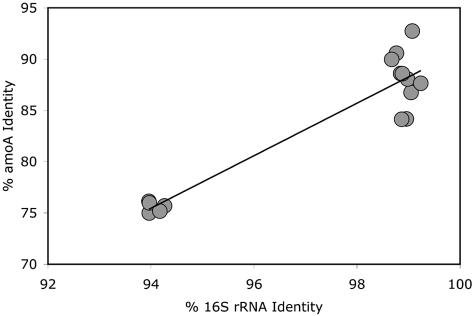
An analysis of πB values (normalized to sequence fragment length to allow for comparison to cultivated strains) for amoA and 16S rRNA genes found a strong positive correlation (r2 = 0.88; F ratio = 95.73; P < 0.0001) (Fig. 3), although the data are bimodal and rely heavily on comparisons to the October 99-M sample. The absence of data points in the upper-left or lower-right portion of the plot suggests that the two sequence data sets were sampled from overlapping portions of the same AOB community. Although it is impossible to associate functional and ribosomal genes directly without cultured representatives, the diversity and similarity values from these environmental samples are consistent with genetic distances for the two genes that would be obtained from a well-sampled population with overlap of membership.
FIG. 3.
Correlation analysis of 16S rRNA (295 bp) and amoA (450 bp) percent sequence identity from MB environmental samples. Each data point represents the pairwise comparison of two environmental samples (e.g., October 99-D compared to April 99-M).
Five studies have made the direct comparison of amoA and 16S rRNA in the same environmental sample, but none provide the number of samples or sequences examined in the present study (3, 8, 15, 24, 47). For example, Caffrey et al. (3) examined betaproteobacterial AOB 16S rRNA and amoA in sediments of Azevedo Pond, a saline tidal pond located directly adjacent to MB in Elkhorn Slough. It is interesting that Azevedo Pond sediment sequences were dominated by N. marina-like sequences, while the adjacent MB water column habitat was dominated by Nitrosospira-like sequences. Most (18 of 21) 16S rRNA sequences from Azevedo Pond were N. marina-like, while the remaining 3 sequences were Nitrosospira-like. In agreement with the 16S rRNA data, 8 of 11 amoA sequences examined from the same sample were N. marina-like, while the remaining sequences clustered with Nitrosospira-like sequences. Comparisons from the literature (3, 8, 15, 24, 47) show general phylogenetic agreement between 16S rRNA and amoA in environmental samples and support the more quantitative findings of this study, suggesting that 16S rRNA and amoA clone libraries sample the same or similar AOB populations. Although amoA allows much greater resolution than 16S rRNA, the general trend of pairwise divergence for amoA and 16S rRNA between cultivated strains is also observed in pairwise comparisons of 16S rRNA and amoA quantitative diversity estimators from environmental samples.
Genetic diversity and biogeochemical function.
To study the interaction of environmental variation, AOB diversity, and biogeochemical function (i.e., nitrification rate), five hypothetical associations were considered. First, the environment, independent of AOB diversity and dynamics, could be driving variation in nitrification rate. Second, the environment could cause changes in AOB that control variation in nitrification rate. Third, the environment could cause changes in the total number or relative activity of AOB that controls variation in nitrification rate. Fourth, AOB could be invariant over time, not responding to the changing environment, suggesting that environmental variation at this scale was not relevant to ecological or functional fluctuation. Fifth, variation could be detected in the environment, AOB diversity, and nitrification rates, but the measured variables would not be correlated. This could be due to a complex interaction of multiple parameters, lacking either a dominant cause-and-effect relationship or lacking adequate signal to resolve the dominant relationship.
Analysis of the 2-year data set of hydrographic and biogeochemical variation did not uncover correlations to nitrification rate, suggesting that physical and/or chemical forcing does not directly the control nitrification rate on the measured time scale (44). We therefore rejected hypothesis one to explain the interactions in our data set. Ward also noted that nitrification rates had less variation than expected compared to variation in other coastal systems or to biological processes measured from the same samples (e.g., ammonium assimilation, chlorophyll, bacterial cell count). This may suggest that the AOB community does not respond to environmental variation or that changes in AOB diversity cause a population response that stabilizes function across a variable environment. AOB strains in culture demonstrate a kinetic response to the experimental addition of ammonia (4, 13, 42), but this type of response is typically not observed with natural AOB assemblages (26, 41, 45). AOB population dynamics may explain the lack of a kinetic response in natural assemblages to experimental ammonia increases, resulting in a stabilized nitrification rate over variable conditions.
Temporal stability of ecosystem function as a result of dynamic population structure or activity has been predicted by theory (16, 18, 22, 33), but it is difficult to study experimentally in systems that change population structure slowly (e.g., many eukaryotes). In populations with long generation times, it is more straightforward to measure the effects of total diversity, changes in activity, or spatial niche differentiation rather than dynamic structuring due to temporal variation. Stabilized function across a temporally variable environment should be maximized by a population with high turnover or short generation time (33), such as bacteria. It is important to note that the dynamic component is not total species richness but instead population structure. Our present sampling of AOB populations measures changes in numerically dominant sequences rather than total species (or sequence) richness.
Small but significant genetic differentiation was detected by AMOVA in AOB populations from MB (Tables 3 and 4 and Fig. 1 and 2), with approximately half of the variation only occurring between samples. A Monte Carlo-based test for nonrandom sequence assemblages showed that in most cases (Tables 3 and 4) samples were significantly different from a random distribution of sequences. Significant variation was detected in both the depth profile and temporal samples. Therefore, we can reject hypothesis four, that no variation was present in AOB populations.
Hypotheses two, three, and five all expect dynamic AOB populations to be detected. A Mantel test (35) was used to examine the correlation of Fst values (for both 16S rRNA and AmoA genes) and pairwise differences in the rate of nitrification, but no correlation was found (data not shown). This finding does not indicate a lack of ecological interaction of AOB dynamics and biogeochemical function. It may be that the variation in AOB populations is responsible for maintaining the relatively stable nitrification rates, while the other biological processes examined were substantially more variable (44).
If AOB population dynamics do stabilize function, a correlation between environmental variation and AOB dynamics would be expected to exist (Hypotheses two and three). Hypothesis five expects no correlations among environment, AOB populations, or nitrification rates. Additional Mantel tests examining AOB Fst values together with temperature, salinity, bacterial abundance, nitrite, and nitrate (data not shown) did not yield a significant correlation between environmental variation and AOB population dynamics. Hypothesis three, which expects a correlation between environmental variation and changes in the abundance or activity of AOB, could not be evaluated due to the lack of data on total AOB abundance or differential expression patterns. We are unable to reject hypothesis three or five. Although significant variation was detected in AOB communities, none of the environmental parameters measured can be implicated in controlling the patterns of variation in AOB. It also remains unclear whether AOB population dynamics indicate that changes in the total number of AOB and changes in AOB expression dynamics may act to stabilize nitrification rates or whether no clear pattern links environmental variation, AOB dynamics, and nitrification rates. Additional complications are due to the incomplete information on gammaproteobacterial AOB, which may at times contribute significantly to ammonia oxidation in this environment. The solution to these problems is to increase the number of samples being examined, include mRNA-based assays, and examine samples which have a greater range in the rates of nitrification. We are presently investigating the use of mRNA microarrays and experimentally manipulated systems to further explore these interactions.
Conclusions.
AOB populations from Monterey Bay were dominated by Nitrosospira-like sequences, while AOB populations in the adjacent Elkhorn Slough were dominated by N. marina-like sequences. The MB samples with the highest degree of genetic overlap were separated in time. In contrast, samples with no temporal separation but small vertical separation had comparatively less genetic overlap. AmoA cluster B (Fig. 2) represents novel gene sequences that are phylogenetically distinct from any previously identified AMO sequences. This study is consistent with recent AOB environmental studies that show that the majority of marine AmoA or AOB 16S rRNA sequences are phylogenetically distinct from any cultured strain. AOB 16S rRNA types identified by Hollibaugh et al. (9) as dominant polar types may indeed represent dominant marine AOB. This reinforces the need for novel approaches and additional effort to culture AOB from various environments and provides a number of target groups for culture work.
No correlation between changes in AOB diversity and composition and changes in nitrification rates were found. It appears that, within the nutrient and hydrographic regimes of Monterey Bay, AOB population structure and function are either not tightly linked, are controlled by changes in AOB abundance and expressional dynamics, or lack sufficient variation to discern their relationship. Correlations among AOB community structure and environmental variables and function would be expected to be stronger in environments with steeper gradients and greater temporal variability.
Acknowledgments
We thank Mary Hogan, Neil Harrington, Jane Caffrey, and Darryl Martino for help in the collection of samples and Neil Harrington and Jane Caffrey for technical assistance with biogeochemical measurements.
This work was supported by the NSF (OCE-9896240). G.D.O. received stipend support from an NSF grant to B.B.W. (OCE-9981482).
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1999. Short protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 2.Bano, N., and J. T. Hollibaugh. 2000. Diversity and distribution of DNA sequences with affinity to ammonia-oxidizing bacteria of the β-subdivision of the class Proteobacteria in the Arctic Ocean. Appl. Environ. Microbiol. 66:1960-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caffrey, J. M., N. Harrington, I. Solem, and B. B. Ward. 2003. Biogeochemical processes in a small California estuary: 2. Nitrification activity community structure and role in nitrogen budgets. Mar. Ecol. Prog. Ser. 248:17-40. [Google Scholar]
- 4.Carlucci, A. F., and J. D. H. Strickland. 1968. The isolation, purification and some kinetic studies of marine nitrifying bacteria. J. Exp. Mar. Biol. Ecol. 2:156-166. [Google Scholar]
- 5.Excoffier, L., P. Smouse, and J. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 131:479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis, C. A., G. D. O'Mullan, and B. B. Ward. 2003. Diversity of ammonia monoxygenase (amoA) genes across environmental gradients in Chesapeake Bay sediments. Geobiology 1:129-140. [Google Scholar]
- 7.Freitag, T. E., and J. I. Prosser. 2004. Differences between betaproteobacterial ammonia-oxidizing communities in marine sediments and those in overlying water. Appl. Environ. Microbiol. 70:3789-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gieseke, A., U. Purkhold, M. Wagner, R. Amann, and A. Schramm. 2001. Community structure and activity of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Head, I. M., W. D. Hiorns, T. M. Embley, A. J. McCarthy, and J. R. Saunders. 1993. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J. Gen. Microbiol. 139:1147-1153. [DOI] [PubMed] [Google Scholar]
- 10.Hollibaugh, J. T., N. Bano, and H. W. Ducklow. 2002. Widespread distribution in polar oceans of a 16S rRNA gene sequence with affinity to Nitrosospira-like ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 68:1478-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper, A. B., T. Vannelli, D. J. Bergmann, and D. M. Arciero. 1997. Enzymology of the oxidation of ammonia to nitrite by bacteria. Antonie van Leeuwenhoek 71:59-67. [DOI] [PubMed] [Google Scholar]
- 12.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 13.Jones, R., and R. Y. Morita. 1983. Methane oxidation by Nitrosococcus oceanus and Nitrosomonas europaea. Appl. Environ. Microbiol. 45:401-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalchuk, G. A., J. R. Stephen, W. DeBoer, J. I. Prosser, T. M. Embly, and J. W. Woldendorp. 1997. Analysis of β-Proteobacteria ammonia oxidizing bacteria in coastal sand dunes using denaturing gradient gel electrophoresis and sequencing of PCR amplified 16S rRNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowalchuk, G. A., A. W. Stienstra, G. H. J. Heilig, J. R. Stephen, and J. W. Woldendorp. 2000. Changes in the community structure of ammonia-oxidizing bacteria during secondary succession of calcareous grasslands. Environ. Microbiol. 2:99-110. [DOI] [PubMed] [Google Scholar]
- 16.Lawton, J. H., and V. K. Brown. 1993. Redundancy in ecosystems, p. 255-270. In E. Schulze and H. A. Mooney (ed.), Biodiversity and ecosystem function. Springer-Verlag, Berlin, Germany.
- 17.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risks of gene amplification by PCR as determined by 16S rRNA analysis of a mixed-culture of a strict barophilic bacteria. Microb. Ecol. 21:191-198. [DOI] [PubMed] [Google Scholar]
- 18.Loreau, M., S. Naeem, P. Inchausti, J. Bengtsson, J. P. Grimes, A. Hector, D. U. Hooper, M. A. Huston, D. Raffaelli, B. Schmid, D. Tilman, and D. A. Wardle. 2001. Biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294:804-808. [DOI] [PubMed] [Google Scholar]
- 19.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCaig, A. E., C. J. Phillips, J. R. Stephen, G. A. Kowalchuk, S. M. Harvey, R. A. Herbert, T. M. Embley, and J. I. Prosser. 1999. Nitrogen cycling and community structure of proteobacterial β-subgroup ammonia-oxidizing bacteria within polluted marine fish farm sediments. Appl. Environ. Microbiol. 65:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McTavish, H., J. A. Fuchs, and A. B. Hooper. 1993. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J. Bacteriol. 175:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naeem, S. 1998. Species redundancy and ecosystem reliability. Cons. Biol. 12:39-45. [Google Scholar]
- 23.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 25.Nold, S. C., J. Zhou, A. H. Devol, and J. M. Tiedje. 2000. Pacific northwest marine sediments contain ammonia-oxidizing bacteria in the beta subdivision of the proteobacteria. Appl. Environ. Microbiol. 66:4532-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson, R. J. 1981. 15N tracer studies of the primary nitrite maximum. J. Mar. Res. 39:203-226. [Google Scholar]
- 27.Phillips, C. J., Z. Smith, T. M. Embley, and J. I. Prosser. 1999. Phylogenetic differences between particle associated and planktonic ammonia-oxidizing bacteria of the β-subdivision of the class proteobacteria in the northwestern Mediterranean Sea. Appl. Environ. Microbiol. 65:779-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, J., B. S. Weir, and C. C. Cockerham. 1983. Estimation for the coancestry coefficent: basis for a short-term genetic distance. Genetics 105:767-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rotthauwe, J.-H., K.-P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.SAS Institute. 2002. JMP user's guide. SAS Institute, Cary, N.C.
- 32.Schneider, S., D. Roessli, and L. Excoffier. 2000. Arlequin ver. 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland.
- 33.Schwartz, M. W., C. A. Brigham, J. D., Hoeksema, K. G. Lyons, M. H. Mills, and P. J. van Mantgem. 2000. Linking biodiversity to ecosystem function: implications for conservation ecology. Oecologia 122:297-305. [DOI] [PubMed] [Google Scholar]
- 34.Seitzinger, S. P. 1988. Denitrification in freshwater and coastal marine ecosystems: ecological and geochemical significance. Limnol. Oceanogr. 33:702-724. [Google Scholar]
- 35.Smouse, P. E., J. C. Long, and R. R. Sokal. 1986. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 35:627-632. [Google Scholar]
- 36.Stephen, J. R., A. E. McCaig, Z. Smith, J. I. Prosser, and T. M. Embley. 1996. Molecular diversity of soil and marine 16S rRNA sequences related to β-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 64:4147-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strickland, J. D. H., and T. R. Parsons. 1972. A practical handbook of seawater analysis, 2nd edition, bulletin 167. Fisheries Research Board of Canada, Ottawa, Canada.
- 38.Teske, A., P. Sigalevich, Y. Cohen, and G. Muyzer. 1996. Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure cultures. Appl. Environ. Microbiol. 62:4210-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voytek, M. A., and B. B. Ward. 1995. Detection of ammonium-oxidizing bacteria of the beta-subdivision proteobacteria in aquatic samples using the polymerase chain reaction. Appl. Environ. Microbiol. 61:1441-1450. [Google Scholar]
- 40.Ward, B. B. 1982. Oceanic distribution of ammonium-oxidizing bacteria determined by immunofluorescent assay. J. Mar. Res. 40:1155-1172. [Google Scholar]
- 41.Ward, B. B. 1987. Nitrogen transformations in the Southern California Bight. Deep-Sea Res. 34:785-805. [Google Scholar]
- 42.Ward, B. B. 1990. Kinetics of ammonia oxidation by a marine nitrifying bacterium: methane as a substrate analogue. Microb. Ecol. 19:211-225. [DOI] [PubMed] [Google Scholar]
- 43.Ward, B. B. 2000. Nitrification and the marine nitrogen cycle, p. 427-453. In D. L. Kirchman (ed.), Microbial ecology of the oceans. John Wiley and Sons, New York, N.Y.
- 44.Ward, B. B. Temporal variability in nitrification rates and related biogeochemical factors in Monterey Bay, CA. Mar. Ecol. Prog. Ser., in press.
- 45.Ward, B. B., and K. Kilpatrick. 1990. Relationship between substrate concentration and oxidation of ammonium and methane in a stratified water column. Cont. Shelf Res. 10:1193-1208. [Google Scholar]
- 46.Ward, B. B., and G. D. O'Mullan. 2002. Worldwide distribution of Nitrosococcus oceani, a marine ammonia-oxidizing gamma-proteobacterium, detected by PCR and sequencing of 16S rRNA and amoA genes. Appl. Environ. Microbiol. 68:4153-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster, G., T. M. Embley, and J. I. Prosser. 2002. Grassland management regimes reduce small-scale heterogeneity and species diversity of proteobacterial ammonia oxidizer populations. Appl. Environ. Microbiol. 68:20-30. [DOI] [PMC free article] [PubMed] [Google Scholar]