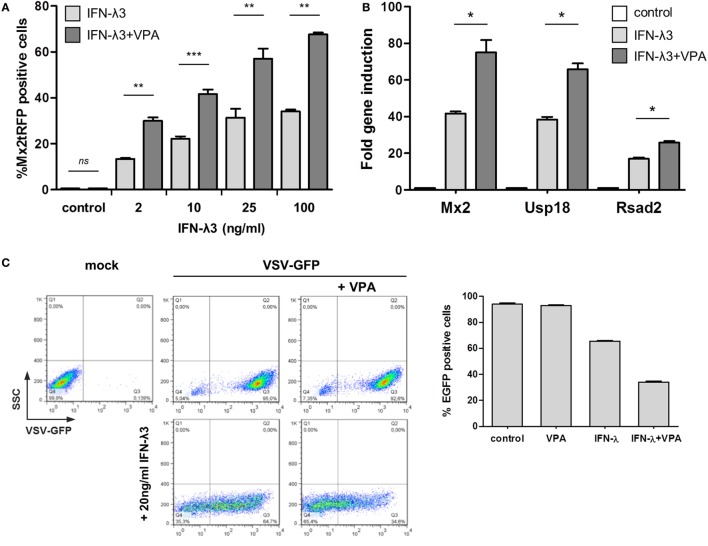
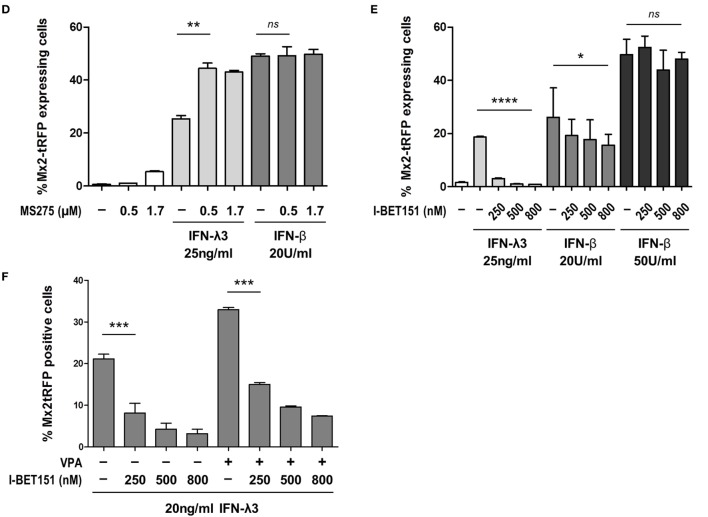
Figure 3.
Differential sensitivity of type I and type III interferons (IFNs) to inhibition of histone deacetylase and BRD3/4. (A) Intestinal epithelial cell lines (IECs) harboring Mx2tRFP were stimulated with increasing concentrations of IFN-λ3 in the absence or presence of 750 µM valproic acid (VPA). Frequency of Mx2tRFP expression was determined by flow cytometry after 24 h (n = 3–6, mean ± SEM). P values were calculated by paired t-test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). (B) IECs were either untreated or treated with 10 ng/ml IFN-λ3 in the absence or presence of 750 µM VPA for 16 h. RNA was isolated, and qRT-PCR was used to determine the expression of Mx2, Usp18, and Rsad2. IFN-stimulated gene expression was normalized to β-Actin (n = 3, mean ± SEM). *P ≤ 0.05 by Mann–Whitney U test. (C) IECs were stimulated with 20 ng/ml IFN-λ3 in the absence or presence of 750 µM VPA for 20 h (lower panel). Control cells were not treated with IFN-λ (upper panel). Cells were infected with vesicular stomatitis virus-GFP (MOI 1) for 1 h and analyzed for eGFP expression by flow cytometry 8 h post-infection. Representative FACS dot plots show the percentage of eGFP-expressing cells. Graph represents mean eGFP frequency for each condition (n = 3, mean ± SEM). (D) IECs harboring Mx2tRFP were stimulated with 20 U/ml IFN-β or 25 ng/ml IFN-λ3 in the absence or presence of the indicated concentrations of MS275. Frequency of Mx2tRFP expression was determined by flow cytometry after 24 h (n = 3, mean ± SEM). **P ≤ 0.01 by paired t-test. (E) IECs were stimulated with IFN in the absence or presence of the indicated concentrations of I-BET151. Percentages of Mx2tRFP-positive cells are given (n = 3–9, mean ± SEM). P values were calculated by one-way ANOVA (*P ≤ 0.05, ****P ≤ 0.0001). (F) IECs harboring Mx2tRFP were stimulated with 20 ng/ml IFN-λ3 in the absence or presence of 750 µm VPA. As indicated, different concentrations of I-BET151 were added during stimulation, and flow cytometry was performed after 24 h (n = 3, mean ± SEM). P values were calculated by one-way ANOVA, followed by Tukey’s Multiple Comparison Test (***P ≤ 0.001).