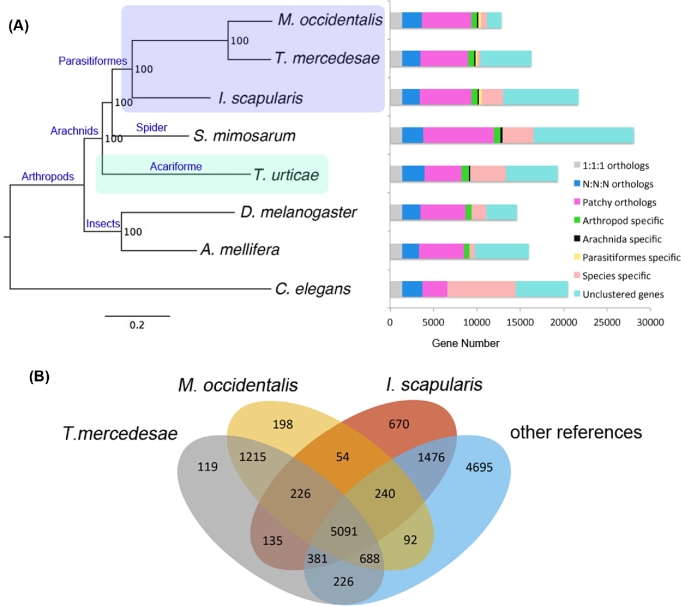
Figure 2:
Comparative genomics. (A) The species phylogeny was built from aligned protein sequences of 926 one-to-one orthologs in Metaseiulus occidentalis, Tropilaelaps mercedesae, Ixodes scapularis, Stegodyphus mimosarum, Tetranychus urticae, Drosophila melanogaster, Apis mellifera, and Caenorhabditis elegans using a maximum likelihood method. The tree was rooted with C. elegans. All nodes showed 100% bootstrap support. Protein-coding genes were classified into the different categories. 1:1:1 orthologs and N:N:N orthologs represent the common orthologs with the same copy numbers and different copy numbers, respectively. Patchy orthologs are shared between more than one but not all species (excluding those in the previous categories). Unclustered genes represent genes which were not classified into orthology cluster. Other categories include arthropod-, Arachnida-, Parasitiformes-, and species-specific genes. C. elegans was used as the outgroup for classification of the protein-coding genes. (B) The number of gene families shared between T. mercedesae, M. occidentalis, I. scapularis, and other reference species (S. mimosarum, T. urticae, D. melanogaster, A. mellifera, and C. elegans) by orthoMCL classification algorithm.