Algorithm 1.
2SNV Algorithm
| Procedure 1: constructing the consensus haplotype for all reads: |
Initialize the set of all clusters with a single cluster with all reads
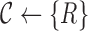
|
| For each position i find allele of highest frequency ai |
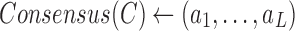
|
| Procedure 2: partitioning reads into simple clusters |
| while not all clusters are simple do |
for each non-simple cluster
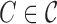 do do
|
| if no pair SNVs is linked according to (2–4) then |
| Regard C as a simple cluster |
| else |
| Find a pair of linked SNVs I2 and J2 minimizing (3) |
Find the set C1 of all reads with the 2-haplotype 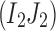
|
Find the consensus 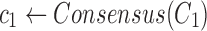
|
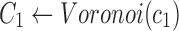
|
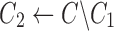 , , 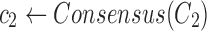
|
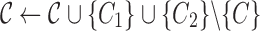
|
| Procedure 3: estimating frequencies of the consensuses of simple clusters |
Run kGEM algorithm for the set of haplotypes 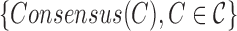 . . |
