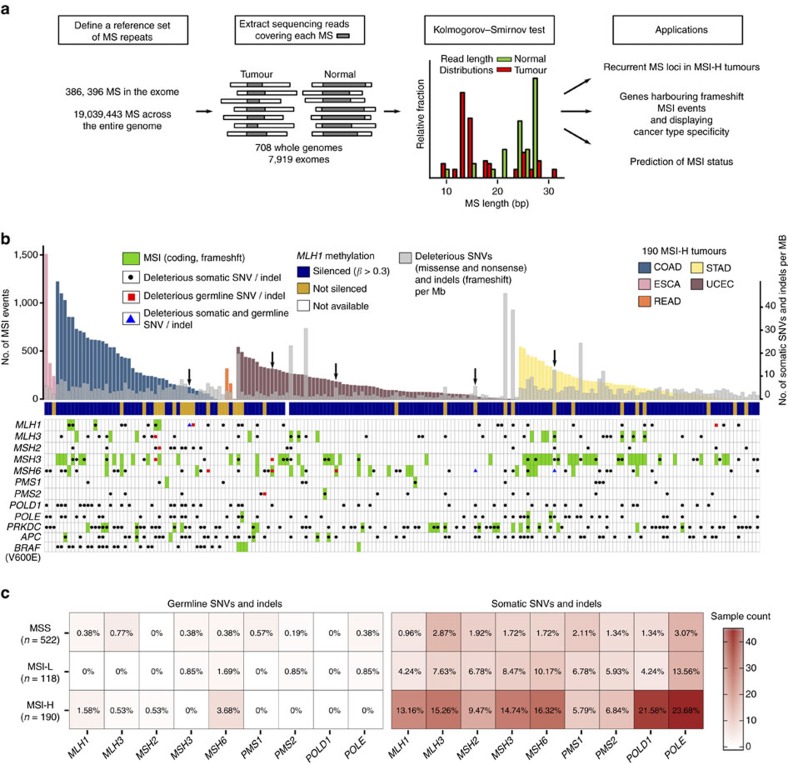
Figure 1. Schematic overview of the MSI calling pipeline.
(a) A reference set of exonic and genome-wide MS repeats was assembled from the human reference genome hg19. The sequencing reads spanning each MS repeat and at least 2 base pairs at each flanking side were extracted from the tumour and normal BAM files. This process was repeated for all MS repeats in the reference sets across all pairs of matched normal-tumour samples. The Kolmogorov–Smirnov test was used to evaluate whether the read length distributions from the normal and tumour samples differed significantly (FDR<0.05). The exonic and genome-wide MSI calls served to identify MS loci recurrently altered by MSI in MSI-H tumours, discover frequent frameshift mutations and to predict MSI status. (b) Landscape of somatic MSI in MSI-H tumours. MSI events (frameshift and in-frame), deleterious SNV (missense, nonsense and splice site) and indel (frameshift) rates in 190 MSI-H exomes. Samples harbouring hypermethylation of the MLH1 promoter are denoted by blue squares. Deleterious germline and somatic mutations (that is, missense, nonsense, splice site and frameshift) are depicted in black and red, respectively, whereas frameshfit MSI events are shown in green. Black arrows mark patients with germline and somatic mutations in MMR genes. (c) Germline and somatic mutations in MMR genes, POLE and POLD1 in MSS, MSI-L and MSI-H tumours. The heatmap and the cell labels report the number and percentage of samples in each category harbouring mutations, respectively.