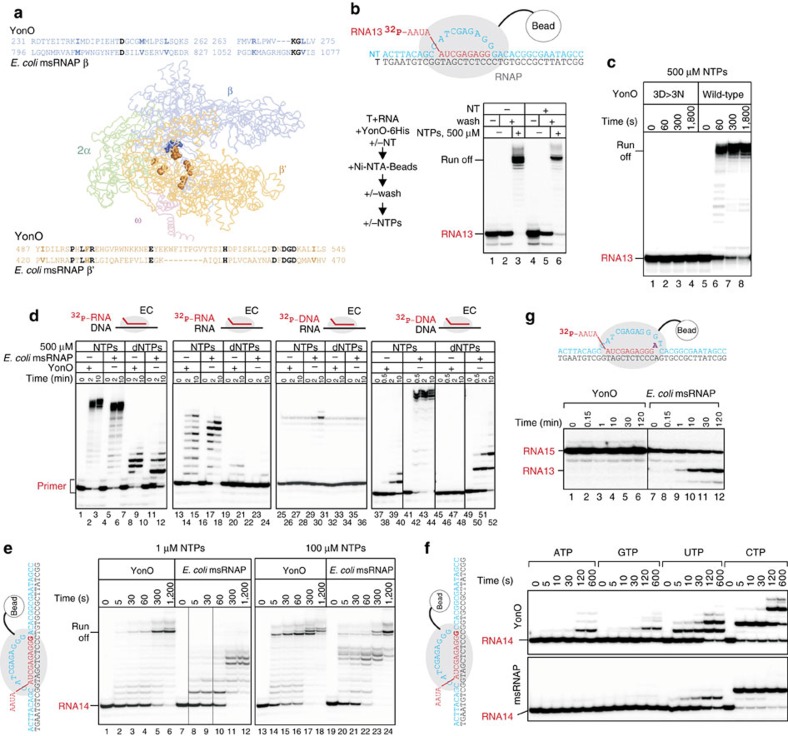
Figure 1. YonO is an RNAP.
(a) Detectible homology of YonO and msRNAP is marked in the alignment (identical amino acids—black, conserved substitutions—bold). Identical amino acids are shown on the crystal structure of E. coli msRNAP (pdbID 4IGC) as spheres. (b) YonO forms stable active elongation complexes. Scheme of the experiment and the sequences of nucleic acids used are shown next to the gel (here and after, for sequences, see Supplementary Table 2). Partial destruction of ECs upon addition of access of the non-template DNA strand (lane 5) is also commonly observed for msRNAP. (c) RNA extension in ECs (as in panel (b)) formed by wild-type YonO and mutant YonO carrying asparagine substitutions of the aspartate triad homologous to the absolutely conserved catalytic aspartate triad of msRNAPs. (d) Specificity of YonO and msRNAP to RNA versus DNA as primers and templates (see Supplementary Table 2 for sequences), and NTP versus dNTP as substrates. A higher molecular weight band in the third panel that coincides with the extension product is a contaminant in the preparation of the DNA primer. (e) Kinetics of DNA-dependent RNA polymerization by YonO and msRNAP in the presence of all NTPs. RNA in the EC was labelled at the 3′ end by incorporation of α-[32P]GMP, shown in bold in the scheme next to the gel. (f) YonO is more error prone than msRNAPs. Kinetics of misincorporation by YonO and E. coli msRNAP. RNA was labelled as in panel (e). Note that RNAs of different sequences are well resolved in this Urea-PAGE excluding the possibility that the extension is caused by the contamination with correct NTPs. (g) Kinetics of RNA hydrolysis by YonO and msRNAP (see also Supplementary Fig. 1f).