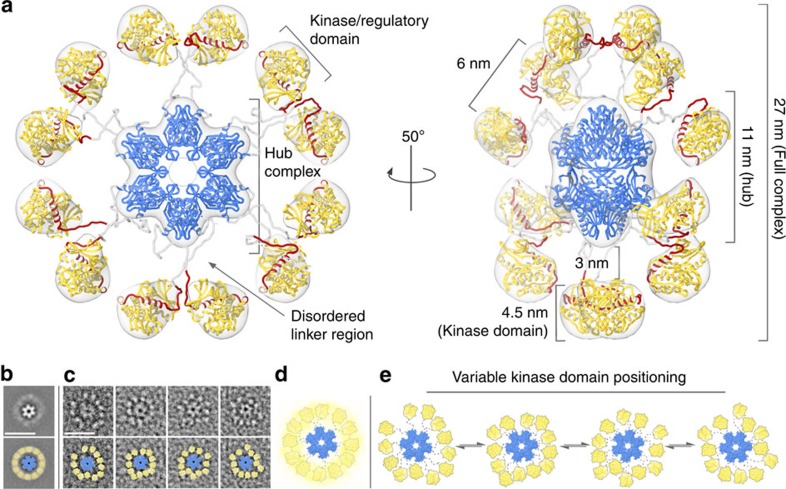
Figure 2. 3D-reconstruction and pseudo-atomic model of the CaMKIIα holoenzyme.
(a) 3D-reconstruction of the CaMKIIα holoenzyme (grey transparent) refined to ∼20 Å resolution. A pseudo-atomic model of the dodecameric holoenzyme (coloured as in Fig. 1a) was constructed using previously determined crystal structures corresponding to the dodecameric CaMKIIα hub complex (blue ribbon, PDBID 5IG3 (ref. 22); residues 345–472) and isolated kinase/regulatory domain (yellow/red ribbon, PDBID 2VZ6 (ref. 21); residues 13–300). Residues 301–344 were modelled as disordered linkers connecting each kinase domain to the nearest hub domain. Residues 274–314 correspond to the regulatory domain (red), containing a proximal auto-inhibitory segment and distal calmodulin-binding site. (b,c) Unmasked 2D class average and single particle images (as shown in Fig. 1) with crystal structures of the hub complex (blue surface) and kinase/regulatory domain (yellow surface) fit into the EM densities. A yellow halo in b represents the diffuse positioning of kinase domains in the 2D class average. Scale bars=25 nm. (d,e) Enlarged view of the fit domains in b,c illustrating the proposed structural equilibrium and variable kinase domain arrangements observed in single particle images. Flexible linkers connecting individual kinase domains to the central hub complex are represented as grey dotted lines.