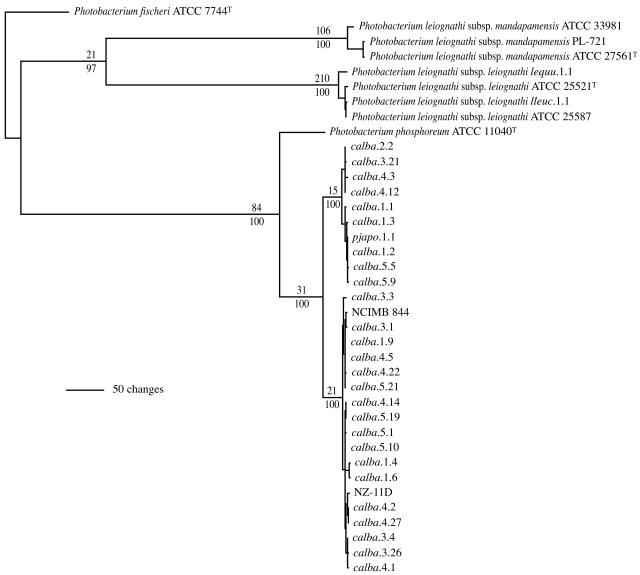
FIG. 5.
Phylogram representing 1 of 18 equally most parsimonious hypotheses resulting from analysis of luxAB(F)E gene sequences of Photobacterium strains (946 informative characters; length = 1,444; CI = 0.866; RI = 0.951). The 18 hypotheses differ only in the resolution within P. leiognathi subsp. leiognathi, and within (but not between) the two subgroups comprising the C. albatrossis strains. The analysis includes C. albatrossis strains from Fig. 1 and 2 and additional strains identified by genomic profiling as genomotypically distinct. Excluded from the analysis, for computational simplicity and visual clarity, were the several strains found to be identical in luxABFE sequence. The strain(s) with luxABFE sequence identical to those included in the analysis were as follows: calba.1.2 = (same sequence as) calba.2.1 and calba.4.4; calba.1.9 = calba.3.2, calba.3.6, calba.3.10, calba.3.12, calba.3.19 and calba.3.25; calba.2.2 = calba.3.7, calba.3.8, calba.4.6, calba.4.12 and calba.4.15; calba.3.3 = calba.3.9; calba.3.4 = calba.3.5 and calba.3.17; calba.3.26 = calba.3.27 and calba.3.28; calba.4.14 = calba.5.3; and calba.5.5 = calba.5.14. Numbers above and below the branches and abbreviations are as in Fig. 1.